Molecular Genetics
tRNA Model
Protein
Ribosomes
Polysomes
-structure of multiple ribosomes attached to same mRNA
-production of proteins at faster rate
Free
Synthesis of proteins used within the cell.
Proteins translated on free ribosomes function in mitochondria, chloroplasts, cytoplasm or nucleus of cell.
Bound
Synthesis of proteins secreted out of cell. (by lysosomes)
Proteins functioning within the ER are synthesized on the bound ribosomes attached to the ER.
- Beginning of polypeptide sequence contains signal sequence recognized by SRP
- SRP binds to SRP receptor protein on ER
- This allows the polypeptide chain to enter rough ER as it grows in length
- When translation ends, whole polypeptide is taken into RER
Translation
Synthesis of a protein from an mRNA template strand.
Aminoacyl-tRNA synthetase
Enzyme which pairs corresponding amino acid to tRNA binding site.
Aminoacylation occurs as the enzyme acts as catalyst, charging the tRNA using ATP.
- Termination codon is reached (termination site)
- Release factor binds to A site, recognizing termination site, thus disassembling the translation complex. (Process is over and all parts are reusable)
Elongation (repeated cycle of events)
- Aminoacyl tRNA binds to A site and has special amino acid matching the mRNA codon sequence and tRNA anticodon.
- New amino acid joins the existing initiator tRNA's polypeptide chain by peptide bond
- Original tRNA molecule with the polypeptide chain exits the translation complex through E site (Ready to be RECYCLED)
- The tRNA molecule in the aminoacyl site translocates to the P site where the OG tRNA used to be and completes the cycle with a longer polypeptide chain
- Whole process restarts until termination codon
- mRNA binds to small ribosomal subunit
- Anticodon of initiator tRNA molecule binds to start codon
- Translation complex completed when large ribosomal subunit joins small one
Transcription
Synthesis of mRNA strand from original DNA molecule in preparation for Translation .
Post-Transcriptional Modifications
In Eukaryotes, before being able to be translated into a protein chain, mRNA strands must be processed into their mature state.
This involves removing ( splicing ) any '' introns '' which are non-coding regions which will not be expressed.
Mechanism of Action:
- snRNPs bind to ends of introns
- Intron is forced into a looped form by the formation of a spliceosome (complex of 5 snRNPs)
- Intron is excised (cut)
- The remaining Exons bind back together
Termination
Process:
- Process continues until reaching a Termination Site
- Transcription stops and mRNA strand is released from DNA
Elongation
Process:
- Ribonucleoside Triphosphates line up with opposite complementary base pairs
- Ribonucleotides are added to 3' side (added from 5' to 3' direction)
- The mRNA strand gets longer as more ribonucleotides are synthesized into the strand
Initiation
Process:
- RNA Polymerase attaches to initiation site at promoter region.
- DNA unwinds double helix in preparation for replication
General DNA
Gene Expression
Non-Coding DNA
Other Functions of DNA:
-Regulation of Gene Expression
-Centromeres, Telomeres
-Introns
-Coding for tRNA
-'Jumping Genes'
Regulating Gene Expression
Producing Proteins takes a LOT of ENERGY
--> Proteins are not needed all the time so we need to REGULATE transcription
Transcription Factors: Interact with polymerase at promoter region.
Activator Proteins: Proteins binding to DNA Enhancer sites outside promoter region in order to upregulate gene expression.
Repressor Proteins: Proteins binding to DNA Silencer sites outside promoter region in order to downregulate gene expression.
Methylation
DNA Methylation
Methylation in DNA blocks gene expression by adding a methyl onto a Cytosine base.
The nucleotide is ' shut ' down . Methylation causes one of the X chromosomes to completely shut down thus making it non-functioning.
Methylation is copied/passed down to daughter cells as it does not affect replication.
Histone Methylation
When histones bind to DNA they block all access to RNA polymerase.
Acetylation of histone --> adding acetyl group to histone allows for gene expression (enzyme can access gene)
Deacetylation of histone--> removing acetyl group silences gene
Methylation of histone --> adding methyl group instead of acetyl affects transcription depending on location of histone (good or bad)
DNA Packaging
Packaging: '' Beads on a string '' - 8 core histone proteins Bundled together by linker DNA and nucleosome
Loosely/free floating = chromatin
Tightly packed together (supercoiled) = chromosomes
DNA Structure
Double helix
-Complementary Base Pairs (A, T, C. G)
RNA
-BAse Pairs: (A U G C)
Phosphate Group
Purines
Nitrogenous bases:
Adenine (A)
Guanine (G)
Pyrimidines
Nitrogenous bases:
Cytosine (C)
Thymine (T)
Uracil (U)
Pentose Sugar
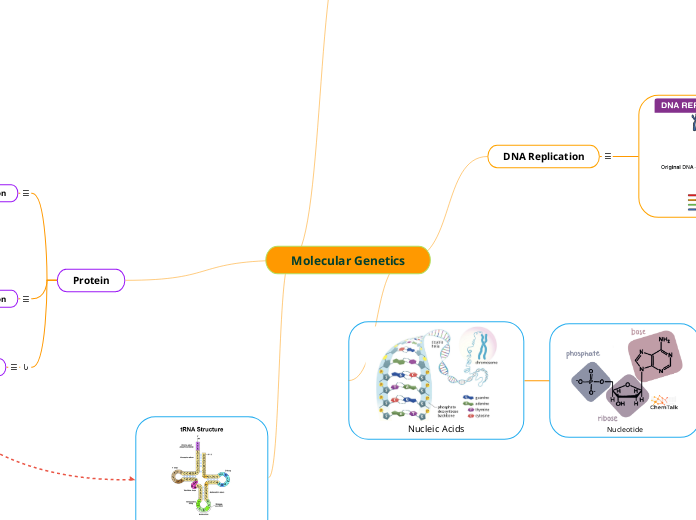
Nucleic Acids
Nucleotide
DNA Replication
Mechanism of Action:
- Helicase Enzyme binds to DNA molecule causing it to unwind from double helix shape
- DNA molecule separates into 2 strands forming a replication fork
- Primase Enzyme binds to the leading strand, adding the RNA bases to form a small RNA strand called primer .
- DNA Polymerase Enzyme binds to the leading strand and begins to move from 5' to 3' end, adding free nucleotide bases.
- Once reaching the next strand, due to the antiparallel nature of both strands, DNA polymerase can only add strands in small chunks to lagging strands called Okazaki fragments.
- Each fragment is started with an RNA primer added on by primase enzyme. Then polymerase adds a few DNA bases to follow primer.
- Exonuclease binds to both strands to remove all RNA primers.
- RNA primers are replaced with new nucleotide bases added by another DNA polymerase enzyme protein.
- DNA Ligase Enzyme seals up the fragments in order to create 2 fully continuous strands.