A multiomics project
DATA
Phenotypic
Clinical
Lab results
Questionnaire
Self reported
Genomics
#samples
14300
Proteomics
#samples
orig 3000
inclusion/exclusion
2000
deficient
<12
399
insufficient
>=12,<20
821
sufficient
>20
655
incl = do not suffer from chronic diseases
exc = anyone with pregnancy
Metabolomics
#samples
orig 3000
inclusion/exclusion
2000
exc = anyone with pregnancy
incl = do not suffer from chronic diseases
AIM
xxxxxx
Objectives
OBJ1
OBJ2
OBJ3
OBJ4
Preliminary work
previously we look at
effects of vitamin D on HDL production
its anti-inflammatory potential
its molecular pathways associated with vitamin D deficiency and dyslipidemia
#samples
n=270
case
~180
deficient
control
~90
sufficient
Gaps
some ppl have higher rate of deciciency compared to others
this means there might be genetic/inherited factors
certain genes linking vitD defeciency with
WORKFLOW 1:
get list of proteins enriched in vitD deficiency from our proteomics study (n=270)
find the genetic variants associated w them
direct
coding region
high impact
it affects protein structure formation
moderate impact
it reduces the effectiveness of a protein
indirect
coding but modifier
it regulates the expression of the protein
non-coding region
it regulates the expression of the protein
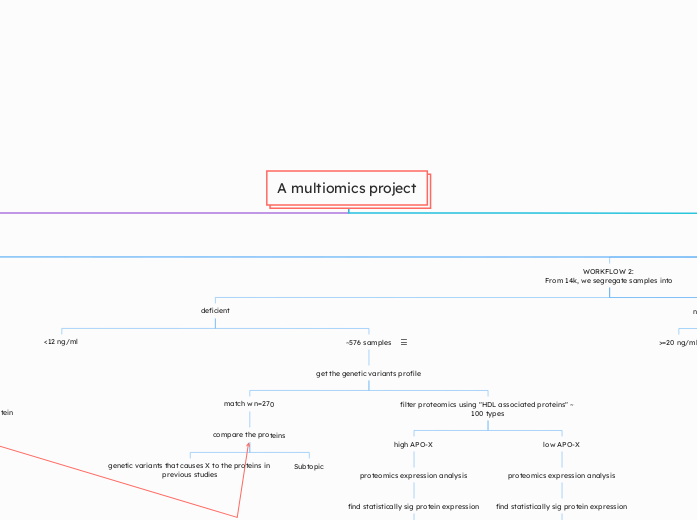
WORKFLOW 2:
From 14k, we segregate samples into
deficient
<12 ng/ml
~576 samples
get the genetic variants profile
match w n=270
compare the proteins
genetic variants that causes X to the proteins in previous studies
Subtopic
filter proteomics using "HDL associated proteins" ~ 100 types
high APO-X
proteomics expression analysis
find statistically sig protein expression
compared genetics w protein exp
HIGH
MODERATE
MODIFIER
gtex
low APO-X
proteomics expression analysis
find statistically sig protein expression
compared genetics w protein exp
HIGH
MODERATE
MODIFIER
gtex
normal/sufficient
>=20 ng/ml
~1049 samples
get the genetic variants profile
insufficient
>12ng/ml, < 20 ng/ml
~1223 samples
get the genetic variants profile
previous work by others
3 dominant gene variants
GC
rs2282679
NADSYN1
rs12785878
CYP2R1
rs10741657
CASE STUDY:
VITAMIN D & CALCIUM
Step1:
Categorize groups - HM
Vit D normal & normal lipid
lipid threshold
cholesterol
triglycerides
LDL
HDL
851
Vit D normal & dyslepedimia
199
Vit D def & Dyslepidimia def
114
Vit D def & normal lipid
463
Step2: Perform proteomics diff expression - RZ
Step3: Check SNPs in the protein statistically diff - RZ
identify impact status
High
affects prot structure
Moderate
affecting effectiveness
Step 5: Check X levels in lab results, questionnaires, EMR - HM
get baseline characteristics
OBJECTIVES
1. Identify genetic mechanism of Vit D defiency
2. Proteomics expression of Vit D deficiency
case study
VitD
deficient
sufficient
insufficient
SLE
Asthma
MS