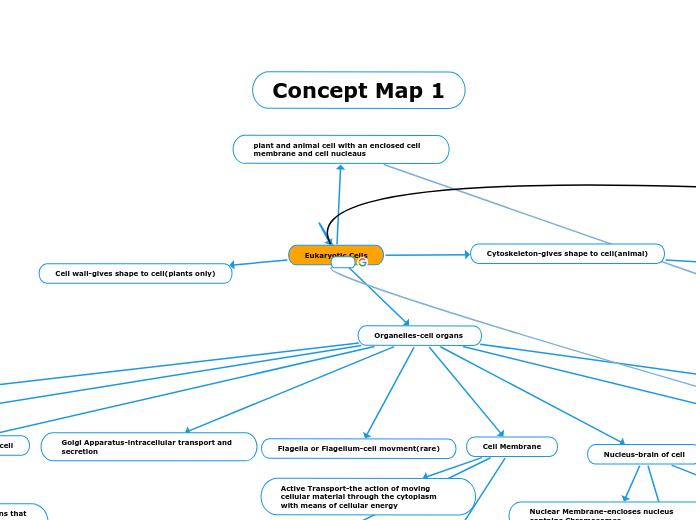
Eukaryotic Cells
Organelles-cell organs
Nucleus-brain of cell
Chromosomes-holder of genetic information
Chromatin-what makes up Chromosomes
DNA-genetic information
Nuclear Membrane-encloses nucleus contains Chromosomes
Nucleolus-a structure surrounding the nucleus during interphase
Ribosomes-made up of RNA, it is responsible for protein synthesis
Proteins-make up Ribosomes
Endoplasmic Reticulum-responsible for things like protein synthesis, protein folding, lipid and steroid synthesis.
Rough ER-ribosomes attach
Smooth ER-ribosomes cannot attach
ribosomes leaving in vesicles from ER to cause protein synthesis and combination with products like carbohydrates and lipids
Golgi Apparatus-intracellular transport and secretion
Vacuole-cavity containing fluid(plants only)
Lysosome-waste management
lipids proteins and one enzyme make up Lysosomes
Mitochondria-powerhouse of the cell
Cellular Respiration-chemical reactions that cause glucose to breakdown into ATP causing cell energy production
ATP-energy in the cell
Chloroplast-photosynthesis(plants only)
Chlorophyll-cellular solar panels
Flagella or Flagellum-cell movment(rare)
Cell Membrane
Active Transport-the action of moving cellular material through the cytoplasm with means of cellular energy
Protein Transport
Channel Protein-water and small ions pass through
Carrier Protein-a protein which has a substance it transports across the cell
Gated Channel Protein-a gate must open for a molecule to pass through
Cytoplasm-jelly like substance filling cell
Vacuoles-space in Cytoplasm
Cytosol-a component of the cytoplasm where organelles and particles are suspended
carbohydrates-found in Golgi Apparatus and Cytoplasm
lipids-found in Golgi Apparatus and Cytoplasm
Cytoskeleton-gives shape to cell(animal)
Microfilaments-the equivalent to muscle contractions
protein
Microtubules-made up of a-tubulin and b-tubulin. also takes part in cell growth and intracellular movement
Diffusion-movement from a high concentration to a power concentration. both solute and solvent molecules move freely
Osmosis-movement from a high concentration to a power concentration. only solvent molecules move freely
Centriole(only active during cell division in animal cells)
Cell wall-gives shape to cell(plants only)
plant and animal cell with an enclosed cell membrane and cell nucleaus
Floating topic
Biomolecules
Molecules created by
living organisms.
Carbohydrates
Simple (Sugars)
Monosaccharides
Pentoses: 5-carbon
Ribose
Deoxyribose
Hexoses: 6-carbon
Glucose
Alpha: OH is below the
ring
Beta: OH is above the ring
Galactose
Fructose
Disaccharides
Sucrose
Lactose
Maltose
Complex
Storage polysaccharides
Glycogen
Alpha glucose & 1-4 and 1-6
glycosidic linkages (lots of branching)
Starch
Amylose
Alpha glucose & 1-4 glycosidic
linkages (no branching)
Amylase
Alpha glucose & 1-4 and 1-6
glycosidic linkages (some branching)
Dextran
Structure polysaccharides
Cellulose
Beta glucose & 1-4 glycosidic
linkages (no branching)
Chitin
Nucleic Acids
DNA
deoxyribose sugar
phosphate
nitrogenous base
Purines
Adenine (A) & Guanine (G)
Pyrimidines
Cytosine (C) & Thymine (T)
RNA
ribose sugar
phosphate
nitrogenous
base
Purines
Adenine (A) & Guanine (G)
Pyrimidines
Cytosine (C) & Uracil (U)
Gene expression
Lipids
Triglycerides
Glycerol
3 Fatty acids
Saturated
No double bonds & H atoms
at every position
Solid at room temp
Unsaturated
One or more
double bonds &
H atoms are notat every position
Liquid at room temp
Cis: H on same side
of double-bonded carbons.
Has a kink
Trans: H on different sides
of double-bonded carbons
Energy storage
Steroids
4 fused rings
Cholesterol
HDL: Carries excess to
liver for excretion (good)
LDL: Carries excess to
blood vessels (bad)
Testosterone
Phospholipids
Hydrophobic tail:
Glycerol &
2 Fatty acids
Hydrophilic head:
Phosphate group
Form bilayer in water
Proteins
Amino acids
Main chain: Amino &
carboxyl groups
Side chain:
R group
Polar: Has OH, SH,
or NH groups
Nonpolar: Has H, CH,
or carbon ring
Acidic: Complete
negative charge
Basic: Complete
positive charge
Polypeptide
Primary: Amino acids
connected through peptide
bonds
Secondary: Main chains form
hydrogen bonds
Alpha helix
Beta-pleated sheet
Tertiary: R groups interact
to from 3D shape
Hydrogen bonds
Hydrophobic
interactions
Ionic bonds
Disulfide bond
Quaternary: 2 or more
polypeptides form a
functional protein through
R group interactions
Monomer: 1 PP
Dimer: 2 PPs
Trimer: 3 PPs
Tetramer: 4 PPs
dehydration reaction
hydrolysis
Floating topic
Cell membranes
Function
transport
smaller molecules
facilitated diffusion
passive transport facilitated by proteins
channel proteins are needed
active transport
protein pumps
low->high concentration
ex:sodium-potassium pump
osmosis/diffusion
Isotonic
Hypotonic solution
turgid(normal in a plant cell)
Hypertonic solution
plasmolyzed (plant loses water)
larger molecules
endocytosis
pinocytosis
phagocytosis
receptor-mediated
exocytosis
barrier, support, protection
separates organism from the environment
Structures
are
selectively permeable
made of
phospholipid bilayer
hydrophobic tail (fatty acid chain) and hydrophilic head (polar) which help control membrane fluidity
proteins
Ion channels
Un-gated (always open)
Gated (stretch-gated, ligand-gated, voltage-gated)
sodium potassium-pump, potassium channel, sodium channel
Transmembrane proteins have an extracellular and cytoplasmic side
transport, enzymatic activity, cell-cell recognition, signal transduction, intercellular joining, and attachment to the cytoskeleton and extracellular matrix (ECM)
Floating topic
Floating topic
Floating topic
Floating topic
Prokaryotes
Nutrition Modes
Autotrophs
Photoautorophs
Light
Chemoautoroph
Inorganic chemicals
Heterotrophs
Photoheterotroph
Chemoheterotroph
Types
Bacteria
Archaea
Types
Extreme halophiles
Extreme thermophiles
Methanogens
Common Structures
Plasma membrane
Specific structures
Transport of molecules
Factors of fluidity
Temperature
Types of fatty acids present
Cholesterol
Fimbriae and pili
Cell wall
Peptidoglycan
Cytoplasm
Ribsomes
Flagella
Capsules and slime layers
Gas vacuoles
Endospore
Nucleoid
Materials Present
Bacterial Genome
RNA
Proteins
Metabolism
Obligate aerobes
Obligate anaerobes
Facultative anaerobes
Concept Map 1
Signal Transduction
Amplification of signals and
coordination/regulation of
cellular response
Reception
Ligand (signaling molecule) binds
to membrane receptor (e.g., GPCR)
GCPR adds GTP to
G protein, which then activates
membrane enzyme
Common relay molecule:
Cyclic AMP
ATP using the enzyme
Adenylyl cyclase
AMP after it activates
the next step. Converted by
phosphodiesterase (PDE).
Transduction
Phosphorylation cascade
Activation of relay molecule
(small, water-soluble molecule/ion),
triggered by reception of ligand
Activation of a protein kinase 1
Activation of protein kinase 2
as protein kinase 1 transfers a
phosphate group to it
Activation of an inactive protein as
protein kinase 2 transfers a phosphate
group to it
Activated protein triggers cellular response
Response
Cellular response is activated
after the transduction pathway
is completed.
Expression of a gene
Photosynthesis
Light Reaction
Photosystem II(P680)
light and water enter the complex
electrons from water are attached to the chlorophyll as protons and O2 are excecated out. light then causes the electrons to jump to an excited state
as the electrons jump to an excited state they are accepted by the primary acceptor
the electrons then go out a electron transport chain causing ATP to be released. the chain consist of plastoquinone (Pq), Cytochrome Complex,
and Plastocyanin (Pc)
Photosystem I(P700)
as the electrons for the ETC enter the chlorophyll and more light exciting them they enter the primary acceptor
this causes another ETC consisting of ferredoxin (Fd) to NADP+ reductase.
NADP(+
)+ 2 H+ binds with NADP+ causing NADPH+H+ to be formed
Cyclic Electron Flow-only used when the cell needs more ATP
Cytochrome Complex to the Plastocyanin creating ATP
plastocyanin then brings its energy to the chlorophyll
electrons get excited to the primary acceptor
electrons are then taken by Ferredoxin back to the Cytochrome Complex
Photophosphorylation: ATP from ETC
is used to pump H+ into thylakoid space.
H+ diffuses down its concentration gradient
through ATP synthase, forming more ATP.
Calvin Cycle
CO2 is introduced due to Rubisco
Phase 1 Carbon Fixation
as CO2 binds with rubisco it creates a short lived intermediate
after the short lived intermediate 3-Phosphoglycerate is made
with the introduction of 6 ATP and excretion of ADP 1,3-Bisphoglycerate is made.
with the introduction of 6 NADPH and the excretion of NADP+P we enter a new phase
NADP+P
Phase 2 Reduction
Glyceraldehyde-3-phosphate
(G3P) is created by phase 1 and is the main sugar used by plants
Phase 3 Regeneration of the CO2 acceptor
3 ATP are introduced cause 3 ADP to leave
this creates Ribulose bisphosphate (RuBP)
CAM Cycle
Temporal separation of steps
C4-Cycle
CO2 enters the mesophyll cell through PEP carboxylase
Oxaloacetate(4C) is formed
malate(4C) is formed
Bundle Sheath cell
CO2
calvin cycle
sugar
vascular tissue
Pyruvate (3C)
PEP (3C)
Spatial separation of steps
Cell Signaling
Physical Contact
Gap Junction (animal cells
Plasmodesmata (plant cells)
Releasing a signal
Local signaling
Paracrine signaling
Synaptic signaling
Long distance signaling
Hormonal Signaling
Receptors
Membrane receptors
includes
G protein linked receptor
signal molecule binds to the GPCR
slight alteration in the shape of GCPR allows for the G protein to bind to it
GDP is replaced with GTP on the G protein
G protein with GTP bound to it is active and it can now activate a nearby enzyme
all of the above steps occur in reception
Tyrosine kinase receptor
Polypeptide on dimerization functions as a kinase
it takes a phosphate group from ATP and adds it to another polypeptide
Ion channel receptor
when a signal molecule binds to the receptor, the gate allows a specific ion like sodium or calcium through the channel in the receptor
movement of ions through these channels may change the voltage across membranes
this would trigger action potential
Intracellular receptors:in cytoplasm & nuclues
steroid hormone aldosterone
target cell that receives the signal molecule
reception
transduction
response
Concept Map 2
Floating topic
Transcription
Initiation
Promoter
TATA box-using for recognition of transcription factors
Transcription Factors-used to RNA polymerase ii can bind in the correct position
RNA Polymerase ii-add RNA nucleotides
RNA polymerase ii and Additional transcription factors bind in order to make a transcription initiation complex.
start point-point of transcription
Elongation
RNA polymerase ii bind to the template strand of DNA 3'-5'
RNA polymerase ii unwinds DNA and then adds RNA nucleotides
the RNA being created resembles the non-template strand of DNA 5'-3'
pre-mRNA strand is now created 5'-3'
Termination
the pre-mRNA is noticed by ribonuclease an amino acid which cuts the pre-mRNA from the DNA
a 5'cap is added to the pre-mRNA
a 3' poly-A tail is added
by polyA polymerase
the pre-mRNA is now ready
to undergo processing
RNA Processing
intron-filler material
exon-material containing suitable nucleotides for protein function
introns are cut out by spliceosome
exons are brought together through spliceosome
mature mRNA is created
and ready for translation
into proteins
Translation
The reading of mRNA codons by tRNA
and its anticodons (attached by aminoacyl tRNA synthatase) to form amino acid chains
direction
similar process
Small ribosomal subunit binds
to the G cap and walks along
the mRNA until it reaches AUG,
the start codon. Initiator tRNA
with anticodon UAC and amino acid
methionine base-pairs with
AUG.
Large ribosomal subunit completes
initiation complex. GTP provides energy
for assembly. The initiator tRNA is in
the P site. The A site is available for
the next tRNA.
Small ribosomal subunit binds an mRNA
and recognizes a specific nucleotide
sequence just upstream of the start codon AUG. Initiator tRNA with anticodon UAC and amino acid formyl methionine base-pairs with AUG.
Elongation
mRNA codon is recognized
by anticodon of incoming
tRNA.
Peptide transferase catalyzes
the formation of a peptide bond
between the amino group of the
new amino acid in the A site and
the carboxyl end of the polypeptide
in the P site.
Ribosome translocates the tRNA
in the A site to the P site. The
empty tRNA in the P site is moved
to the E site, where it is released.
mRNA moves with bound tRNA,
bringing the next codon to the A site.
Termination
The stop codon reaches
the A site. There is no
tRNA that corresponds with
it.
Instead, a release factor sits
in the A site, disassociating the
complex, stopping translation.
Disassociation is driven by GTP.
free ribosomes
Protein Transport
Signal sequences, which
dictate proteins' final
location in the cell
Organelles
Peroxisomes, mitochondria,
chloroplasts (in plants), nucleus
Endomembrane
system
Signal-recognition particle
carries ribosome to endoplasmic
reticulum.
SRP leaves, protein synthesis
continues until finished. Then,
signal peptide is cleaved and the
polypeptide leaves the ribosome
and enters the ER.
Protein travels in a vesicle to the
Golgi apparatus where it is modified.
Modifications include glycosylation,
the addition of carbohydrates to proteins
to make glycoproteins.
Protein can be carried in vesicles to
lysosomes or the plasma membrane
to become a membrane protein
or be secreted.
Gene Regulation
DNA Packing
Histones
First level of packaging involves attaching proteins (histones) to DNA
Made of H2A, H2B, H3, H4 at the core
10 nm fiber
DNA winds around histones to form nucleosome “beads”
30 nm fiber
Interactions between nucleosomes cause the thin fiber to coil or fold into this thicker fiber
300 nm fiber
The 30-nm fiber forms looped domains that attach to proteins
Transcription
signal protein
General
Bring about low levels of transcription (background/basal)
Specific
changes level of transcription
Activators:increases level
Inhibitor: reduces level
In Eukaryotic cells
1) A eukaryotic promoter
commonly includes a TATA
box (a nucleotide sequence
containing TATA) about 25
nucleotides upstream from
the transcriptional start point
2)Several transcription
factors, one recognizing
the TATA box, must bind
to the DNA before RNA
polymerase II can bind in
the correct position and
orientation.
3)Several transcription
factors, one recognizing
the TATA box, must bind
to the DNA before RNA
polymerase II can bind in
the correct position and
orientation.
Prokaryotes
Lac operon
Active
Lactose present, cAMP is present
Lactose present, CAP is active
Lactose present no glucose
Inactive
glucose is present
Concept Map 3
DNA Replication
Fast and accurate
Origin of replication: Sequence of nucleotides that indicates
the beginning of DNA replication
Replication bubble
Bidirectional forks- located at each
end of the replication bubble
Double stranded DNA- with a complimentary, antiparallel structure
Helicase- enzyme
Single-stranded DNA
Single-stranded Binding
Protein (SSBs)- enzyme
Primase-enzyme
Leading strand
Parental DNA
DNA polymerase III
sliding clamp
Daughter strand
5' to 3' direction
Chargaff's rule- states purines and pyrimidine must be paired with one another, specifically the same proportion of
adenine to thymine and guanine to cytosine
Lagging strand
Okazaki fragments
DNA ligase- fills in the gaps left by
removed RNA primase with nucleotides
complimentary to the parent strand
Replication forks- located at each end
of the replication bubble
Topoisomerase- enzyme
Cellular Respiration
Glycolysis
Energy Investment Phase
ATP
Glucose
ADP
Energy Payoff Phase
NADH
Protons
Phosphate
Electrons
NAD+
Protons
Pyruvate
Water/H2O
Pyruvate Oxidation
Pyruvate
c
Citric acid cycle begins
Citrate
Isocitrate
a-ketoglutarate
Succinyl CoA
Succinate
Fulmarate
Malate
Oxaloacetate
Substrate level phosphorylation
Anerobic respiration
Alcohol Fermentation
ADP
NAD+
ATP
NADH
Lactic Acid
ADP
NAD+
ATP
NADH
Oxidative Phosphorylation
Electron Transport Chain
Inner mitochondrial membrane
Chemiosmosis ATP Synthesis
concentration gradient
transport protein - ATP Synthesis