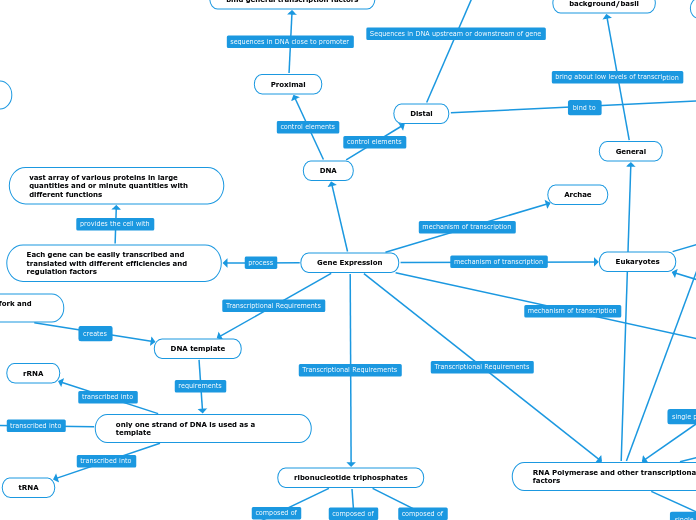
Gene Expression
Each gene can be easily transcribed and translated with different efficiencies and regulation factors
vast array of various proteins in large quantities and or minute quantities with different functions
DNA template
only one strand of DNA is used as a template
rRNA
mRNA
Beta Galactosidase
Permease
Transcetylase
transcription
prokaryotes
transcription in cytoplasm
RNA pol
Eukaryotes
transcription in nucleus
RNA pol II
pre-mRNA
RNA splicing
introns are spliced out
exons remain in final mRNA
5' end cap and 3' poly A tail
occurs downstream from +1
promoter found upstream
Translation/Protein Movement
Ribosomes
mRNA
tRNA
anticodons
Amino Acids
Binding Sites
A Site
P Site
Peptidyl Transferase
E site
codons
Large and Small subunits
Steps in Translation
Eukaryotic
Initiation
Prokaryotic
Initiation
Elongation
Termination
Cytoplasm
Proteins in the Endomembrane System
Secretory Pathway
Polypeptide synthesis on a free ribosome
Endoplasmic Reticulum
Mitochondria
Peroxisomes
Chloroplasts
Nucleus
tRNA
ribonucleotide triphosphates
Nitrogenous base pair
purines
ATP
UTP
GTP
Pyrimides
CTP
Single-ring structure
ribose sugar
sugar phosphate backbone
encodes
phosphate group
RNA Polymerase and other transcriptional factors
three multimeric structures
RNA polymerase II primary one
Orientation and facilitates RNA Polymerase Binding
ATP
elongation
termination
capping
methylation
RNA splicing
poly A tail
site for addition of poly A tail
inactivated by phosphatases
RNA polymerase I synthesized rRNA
RNA polymerase III synthesizes small RNA such as tRNA
General
background/basil
Specific
changes level of transcription
Activators
Repressors
Archae
Eukaryotes
nucleosomes
histones
H1
H2A
histone core
H2B
H3
H4
DNA
Prokaryotes
Binding of RNA to DNA
initiation of transcription
elongation
termination
protein
RNA polymerase
creates phosphodiester bond between NTP and sugar
elongation in 5' to 3' direction
Promoter
Orientation to polymerase
recognized even in helical double stranded form
not transcribed
sigma factor of polymerase recognizes promoter region
Lac operon
Lactose present
repressor binds to operator
Operon is OFF basil
repressor binds to Lactose
Glucose present
Adenylyl cyclase is active
cAMP levels High
CAP is active
Operon is ON
Adenylyl cyclase is inactive
cAMP levels are low
CAP is inactive
Operon is OFF
DNA
Proximal
bind general transcription factors
Distal
Enhancers
in the elongation stage, a polypeptide chain begins as new amino acids are brought to the A site and attached to the previous one. the empty tRNA moves then moves to the P site after the amino acid chain was transferred
no tRNA corresponds to a stop codon, so a release factor in the A site disassociates the complex and stops translating the mRNA (a GTP driven process)
in the ER, translation continues and the signaling peptide is cleaved by an enzyme called Signal Peptidase
Elongation
initiator tRNA scans mRNA for start codon & large subunit joins the translation initiation complex (other proteins called initiation factors may be involved)
Termination
Aminoacyl tRNA Synthetase
Vesicle
Golgi Body
Lysosome
Plasma Membrane
Promotes mRNA polymerase dissociation
with rho factor
requires ATP
short GC rich sequences create mRNA foldiing
without rho factor
DNA Replication
semi conservative proved by Messleson and Stahl experiment
unzipped by Helicase
kept apart by SSB
stress relieved by topoisomerase
replication begins at ORI
5' to 3' direction
leading strand with primase and DNA pol III
lagging strand and DNA pol I
direction of replication goes into fork and ligase seals gaps
Experiments
Griffiths
proved genetic material could be passed through bacteria
Hershey and Chase
protein Sulfur
Found in supernatum
Subtopic
DNA Phosphorus
Settled in pellet