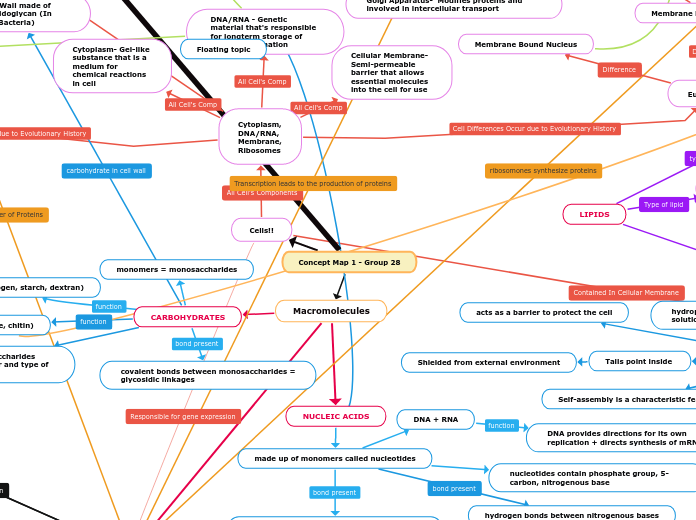
Concept Map 1 - Group 28
Macromolecules
NUCLEIC ACIDS
made up of monomers called nucleotides
nucleotides contain phosphate group, 5-carbon, nitrogenous base
phosphodiester linkage between nucleotides
DNA + RNA
DNA provides directions for its own replication + directs synthesis of mRNA
CARBOHYDRATES
monomers = monosaccharides
covalent bonds between monosaccharides = glycosidic linkages
storage (e.g. glycogen, starch, dextran)
Cells!!
Cytoplasm, DNA/RNA, Membrane, Ribosomes
Eukaryotes
Cell Wall (if any) made of cellulose (or chitin in fungus)
Membrane Bound Organelles
Prokaryotes (Archaea & Bacteria)
Plasmids - genetic structure in a cell that can replicate independently of the chromosomes.
Capsules- Helps prokaryotes cling to each other and to various surfaces in their environment
Cytoplasm- Gel-like substance that is a medium for chemical reactions in cell
DNA/RNA - Genetic material that's responsible for longterm storage of cellular information
Cellular Membrane- Semi-permeable barrier that allows essential molecules into the cell for use
PHOSPHOLIPIDS
A type of lipid, one of the most important components of biochemistry
Major component of the cell membrane
responsible for dynamic membrane fluctuations
Membranes have saturated and unsaturated fatty acids to maintain the proper amount of fluidity
impart selective permeability
they control the movement of molecules across the cell membrane
Hydrophobic behavior ^
Soluble in non polar solvents
Two fatty acids attached to glycerol
glycerol attaches to a phosphate group
Exhibits negative charge within cell
polar hydrophilic head
hydrophilic because of the phosphate group
Non polar hydrophobic tails
The type of hydrocarbon tails effects the plasma membrane fluidity
Bilayer forms by self assembly when contact is made with water
Tails point inside
Shielded from external environment
hydrophilic heads make all contact with the solution
Form because of their amphiphilic characteristics
acts as a barrier to protect the cell
Self-assembly is a characteristic feature
Each phospholipid has a specific transition temperature
goes into a liquid crystalline phase when the temperature is exceeded
phospholipids move rapidly when fluid
movement is reduced as temperature decreases
when this happens the membrane becomes more gel-like.
movement in membranes is regulated by the cholesterol in membranes
linked to signal transductions
& organelle functions
& physiological processes
& human diseases
imperative for cell life
PROTEINS^
Primary
Intramolecular Bonds: Covalent
Uses main chain to form bonds
Polypeptide
Secondary
Beta Plated Sheets
Alpha helices
Tertiary
Folds through interaction of R groups
Non Polar Bonds
Hydrophobic interactions
Acidic
Hydrophilic
Basic
Ion dipole: Complete positive charge with water bond
Forms final 3D shape
Quaternary
Two polypeptides in tertiary level, interacting with R groups
Selective acceleration of chemical reactions
Enzyme
Protection against disease
Subtopic
Storage of amino acids
Transports of substances
Carrier proteins
Coordination of an organism's activities
Response of cell to chemical stimuli
Amino Acids: monomer in proteins
Amino group
Carboxyl group
R Group
Hydrogen
No Membrane Bound Nucleus
Chromosomes are Circular and float around in cytoplasm instead of a nucleus.
Cell Wall made of Peptidoglycan (In Bacteria)
Membrane Bound Nucleus
Ribosomes Function - Synthesizing proteins
Animal
Same Organelles in Both Types of Eukaryotes & Their Functions
Vacuole- Hold important organic materials or hold waste materials inside of cell's
Smooth ER- Synthesizes lipids and detoxifies cells
Rough ER- Ribosomes attached to wall that synthesize proteins
Mitochondria- Responsible for making ATP and is double-membraned and has its own DNA
Plant
Chloroplast- convert light energy into chemical energy via the photosynthetic process, contains plastids, and has its own DNA.
Endosymbiotic Theory- The beginning of eukaryotic cells organelle. Such as mitochondria and plastids, evolved from free-living prokaryotes that were consumed and formed a symbiotic with the cell that ate them. This is supported by the fact that Mitochondria and Chloroplast have their own separate DNA
Central Vacuole- Unlike animal cells, plant cells have a large vacuole in the middle of the cell that holds a large amount of water.
Nucleus- Hold's genetic info, keep DNA integrity, and conduct replication & transcription
Cell Wall- Holds the plants together when exposed to hypotonic solutions that would otherwise burst the cell, making it 'turgid.'
Lysosome- digestive system of the cell, serving both to degrade material taken up from outside the cell and to digest obsolete components of the cell itself.
Vacuole- Animals have small vacuoles that mainly hold organic molecules and are responsible for transport through the plasma membrane.^
Centrosomes- regulates cell motility & adhesion and polarity in interphase,
Golgi Apparatus- Modifies proteins and involved in intercellular transport
Fats (triacylglycerol)
glycerol and fatty acid linked by ester linkages
energy storage
Steroids
contain 4 fused rings
regulates membrane fluidity, component of hormones
LIPIDS
hydrogen bonds between nitrogenous bases
structure (e.g. cellulose, chitin)
structure + function of polysaccharides determined by sugar monomer and type of glycosidic linkage
Concept Map Two- Group 28
Cell Signaling
Local Signaling
Paracrine
Synaptic
Long distance signaling
Hormonal
Membrane Receptors
G Protein linked receptor
Signaling molecule binds to extracellular side of receptor
Receptor activates and changes shape
G protein binds to plasmic side and activates now carrying GTP instead of GDP
Once activated G protein leaves receptor, and binds to enzyme activating it and changing the shape
Tyrosine Kinase receptor
Kinase= adds phosphate groups
Receptor tyrosine kinase proteins (inactive monomers)
Signal molecules bind and form dimer
Phosphate groups are added and the tyrosine kinase receptor is fully activated
Inactive relay proteins bind to receptors and become active
Ion Channel receptor
After Ligand binds the channel opens and specific ions can flow through
Allowing ions to flow through the channel immediately changes concentration inside the cell, and can change the way the cell works
1st messenger
Intracellular Receptors
Signal passes through the plasma membrane
Signal binds to receptor in cytoplasm
Signal-receptor complex enter the nucleus & binds to genes
Cellular Response
1. Reception
2. Transduction
3. Response
Source of ATP
Cellular Respiration
Four Steps In Cellular Respiration
(W/ Presence of Oxygen)
Glycolysis (substrate-level)
Consist of 10 Steps, 5 Energy Investment & 5 Energy Payout Stages
Input's Include- 2 ATP, 2 NAD+, & 1 Glucose
Step One- Glucose turns into Gluc 6- Phosphate w/ the addition of one ATP & Hexokinase
Step Two- Gluc 6-phosphate converts to fructose 6-phosphate w/ phosphoglucoisomerase
Step Three- Fructose 6-phosphate converts to fructose 1,6 bi-phosphate w/ the addition of one ATP & phosphofructokinase
Step Four- Fructose 1,6 bi-phos is cleaved in half by Aldolase which makes it into two three-carb sugars
Step Five- The two three carb sugars both convert to G3P
Step Six- Both G3P's are oxidized, making NAD+ into NADH (electron carrier), free energy is used to add phos groups to G3P
Step Seven- The phos groups of the 1,3 biphosgly is added to ADP to make ATP (2 Total)
Step Eight- The one remaining phos-group of 3-phosgly is relocated by phosphoceromatase
Step Nine- Enolase removes H2O making 2 phos-pyruvate (PEP)
Step Ten- Pyruvate kinase takes phos from PEP thus making 2 ATP & 2 Pyruvate
Quick Overview
Input: 2 ATP, 2 NAD+, & 1 Glucose
Net- 2 ATP, 2 Pyruvate, 2 NADH
Pyruvate Oxidation (substrate-level)
Consist of 4 Steps
Step One- Pyruvate from cytosol (glycolysis) go into the mitochondria
Step Two- The pyruvate gets oxidized & electrons are transferred to NAD+ to NADH
Step Three- Carbon Dioxide is lost, CoA enzyme is added
Acetyl CoA is formed
Quick Overview
Input: 2 Pyruvate, CoA enzyme, 2 NAD+
Output: 2 NADH, 2 Acetyl CoA, 2 CO2
Citric Acid Cycle
(substrate-level)
Consist of 8 Steps
3 Key Steps
1st Step- Acetyl CoA is added to oxaloacetate forming citrate
3rd Step- Isocitrate is oxidized & NAD+ is reduced forming NADH+, CO2 is produced
8th Step- Malate is oxidized, NAD+ converts to NADH, oxloacetate is reformed (restarts cycle)
Quick Overview
Input: 1 Acetyl CoA, 3 NAD+
Output: 1 ATP, 3 NADH, 1 FADH, 2 CO2
Oxidative Phosphorylation
(oxidative)
2 Main Components
Electron Transport Chain (ETC)
Made up of 4 Complexes, I, II, III, & IV
Electrons from NADH & FADH are transported through complexes & release free energy as electrons travel
Free energy is used to pump H+ ions through to the inter-membrane, creating a H+ concentration gradient (complex's also serve as pumps)
Finally, after traveling through complexes & losing energy, electrons are added to oxygen (reducing it)
Chemiosmosis
ATP synthase facilitates diffusion of H+,
b/c of the gradient so H+ is going from high to low (Intermembrane -> Matrix)
Energy is released by the movement of H+ down the gradient, allowing the ATP synthase to add phosphate to ATP to ADP
Quick Overview
Input: NADH & FADH
Output: 26-28 ATP
Cellular Respiration
(W/O Presence of Oxygen)
Alcohol Fermentation
3 Steps
1st- Glycolysis occurs (even w/o oxygen) and 2 Pyruvate form.
2nd- Pyruvate forms 2 Acetaldehyde which reduced to ethanol (2 NAD+ -> NADH)
When reduced, NADH donates electrons so it can be recycled for glycolysis.
Quick Overview
Input: 2 ATP, Glucose, 2 NAD+
Net Output: 2 Ethanol, 2 NADH (Get recycled), 2 ATP, 2 CO2
Lactic Acid Fermentation
3 Steps
Glycolysis occurs (w/o Oxygen)
forms 2 Pyruvate and net 2 ATP
2 Pyruvate us reduced by NADH to form Lactic Acid
NADH gives up electron so it can be recycled and be reused in glycolysis
Quick Overview
Input: 1 Glucose, 2 ATP, 2 NAD+
Net Output: 2 Lactic Acid, 2 NAD+, 2 ATP
PHOTOSYNTHESIS
Location
Photosynthesis occurs within the leaf, within the mesophyll, within the chloroplast.
Mesophyll: The interior tissue of the leaf
Chloroplasts contain chlorophyll: light harvesting pigments
Chlorophyll has chlorophyll a and b
Chlorophyll a contains CH3, chlorophyll b contains CHO
The structure contains a head and a tail.
Porphyrin ring: light-absorbing “head” of molecule, there is a magnesium atom at the center
Hydrocarbon tail: interacts with hydrophobic regions of proteins inside thylakoid membranes of chloroplasts
Consists of PS1 and PS2 along with components of the electron transport chain
H+s are pumped into the thylakoid space, so inside of thylakoid a lot H+ are present
H+ get back out of the thylakoid down their concentration gradient.
The energy from movement down the gradient is used to form ADP
6 CO2 + 18 ATP + 12 NADPH + 12 H2O
C6H12O6 + 18 ADP + 18 Pi + 12 NADP+ 6 O2 + 6 H2O + 12 H+
Plant uses water from soil, light from sun, co2 from atmosphere to form organic compounds
STAGE ONE
Solar energy is converted to chemical energy through light reactions.
In order to provide electrons and protons, H2O is split.
O2 is released as a waste product
NADP+ reduces to NADPH
photophosphorylation generates ATP.
photophosphorylation: The process of generating ATP from ADP and phosphate by means of chemiosmosis, using a proton-motive force generated across the thylakoid membrane of the chloroplast or the membrane of certain prokaryotes during the light reactions of photosynthesis.
STAGE TWO
With the help of the NADPH and ATP produced by the light reactions, The Calvin cycle produces sugar from CO2
Through carbon fixation CO2 is initially incorporated into an organic molecule
In order to reduce CO2, NADPH provides electrons needed, and ATP provides the necessary chemical energy.
The Calvin Cycle
Phase 1: Carbon fixation
Phase 2: Reduction
Phase 3: Regeneration of CO2 acceptor
Takes place in the stroma
Uses ATP and NADPH to convert
CO2 to the sugar G3P
Returns ADP, inorganic phosphate, and NADP+ to the light reactions
Light Reactions
Occur in the thylakoids of the chloroplasts.
Are carried out by molecules in the
thylakoid membranes
Convert light energy to the chemical
energy of ATP and NADPH
Split H2O and release O2 to the
atmosphere
Almost the reversed process of cellular respiration
It's different because Electrons are extracted from water and transferred to CO2. Oxidizing H2O and reducing CO2
As electrons are transferred down the ETC, energy is released which is used to pump H+ against their concentration gradient
Photosystems
Photosystem: reaction-center complex surrounded by light-harvesting complexes.
two kinds of photosystems embedded in the thylakoid membrane
Photosystem II (PS II)-The reaction-center chlorophyll a absorbs at 680
Photosystem I (PS I) The reaction-center chlorophyll a absorbs at 700 nm
Used in cyclic electron flow
The flow of electrons converts to cyclic when there is excess NADPH.
As electrons in PS1 are transferred to Fd, they are recruited to the cytochrome complex and plastocyanin molecules of the electron transport chain.
The movement of electrons leads to formation of ATP by photophosphorylation
Both use in non-cyclic flow of electrons
The light-harvesting complexes transfer the energy of photons to the reaction center.
In the reaction center, chlorophyll a transfers an excited electron to the primary electron acceptor
Light-harvesting complexes: pigment molecules bound to proteins
Reaction-center complex: an association of proteins holding a special pair of chlorophyll a molecules and a primary electron acceptor.
1st messenger
Signal binds to GPCR & activates it
Floating topic
Activated GPCR then binds to G protein, which binds to GTP (activating G protein)
Activated G protein then binds to enzyme called adenylyl cyclase
Adenylyl cyclase then converts ATP into cAMP (2nd messenger)
cAMP activates protein kinase A which causes a cellular response
After cAMP activates PKA, it is converted into AMP by enzyme phosphodiesterase