Molecular Genetics
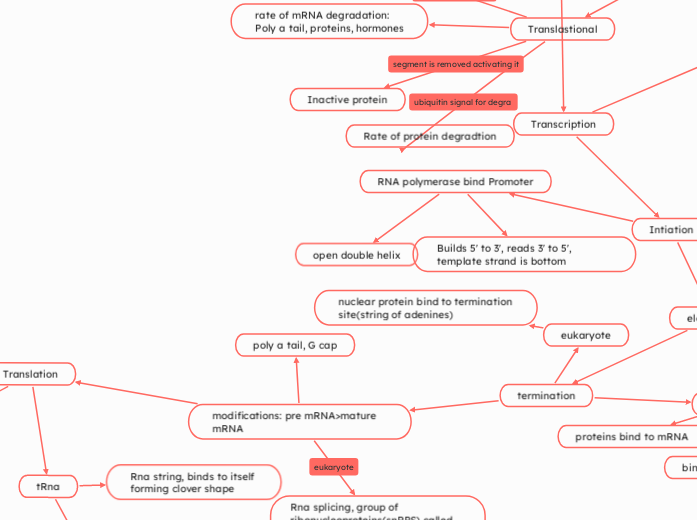
Transcription
Intiation
elongation
termination
modifications: pre mRNA>mature mRNA
Rna splicing, group of ribonucleoproteins(snRPS) called splicesome bring exons close cleave out introns
poly a tail, G cap
Translation
tRna
Rna string, binds to itself
forming clover shape
Aminoacylation, by aminoacyl tRNA synthetase enzyme
Ribosome
intiation
G cap bind to complex, scan for AUG, large subunit is added
Elongation
Peptidly transferase
Ribsome shifts 1 codon, a >p, p>e
Termination
protein release factor binds
prokaryote
binds to itself in hairpin loop
proteins bind to mRNA
eukaryote
nuclear protein bind to termination site(string of adenines)
RNA polymerase bind Promoter
Builds 5' to 3', reads 3' to 5', template strand is bottom
open double helix
Regulation
chromatin accessability
Sepearting tighly wrapped histones
Methylation: Inhibits tanscrip
epigentics
post transcrip
Alternate splicing
masking proteins: inactivate
Translastional
rate of mRNA degradation:
Poly a tail, proteins, hormones
Inactive protein
chem modificationa
Rate of protein degradtion