Cell membrane
Cell
Nucleus
DNA is in the nucleus, and it is bounded by a double membrane.
Nuclear envelope: double
membrane enclosing the
nucleus; perforated by
pores; continuous with ER
Nucleolus: nonmembranous
structure involved in production
of ribosomes; a nucleus has
one or more nucleoli
Chromatin: material consisting
of DNA and proteins; visible in
a dividing cell as individual
condensed chromosomes
This segment of a chromosome
from a nondividing cell shows
two states of coiling of the DNA
(blue) and histone protein (purple)
complex. The thicker form is
sometimes also organized into
long loops.
Prokaryotic cells do not have a nucleus
DNA Structure
Experiments
In 1928, Fredrick Griffith, a British medical officer, was trying to develop a vaccine against pneumonia. He was studying Streptococcus pneumoniae, a bacterium that causes pneumonia in mammals. He had two strains of the bacteria – one pathogenic (disease causing, S strain) and the other nonpathogenic (R strain). The S strain had a smooth surface due to presence of a capsule outside the cell wall, the R strain lacked the capsule. So the presence of capsule made the bacteria pathogenic. So when he injected the S strain into mouse the mouse died, while when he injected it with R strain it lived – as expected. When he injected the mouse with heat killed S strain the mouse lived. But if he added a live R strain with heat killed S components the mouse died. On further analysis, he found that the R strain had changed to S killing the mouse. So something from the S components had entered live R changing the genetic makeup of R to S! What was this component? Are these genes to make capsule present in DNA, RNA, protein or some other molecule? Nobody knew that time…the favored candidate though was protein!
The next experiment done was by Hershey and Chase to determine what was that component that was injected by the bacteriophage inside bacterial cells. As the bacteriophage is only made of two components – DNA and proteins – they labeled DNA of one tube of bacteriophages with radioactive phosphorus (32P) and the proteins of bacteriophages in another tube with radioactive sulfur (35S). They infected bacteria with 35S labeled bacteriophages (shown here as pink) and another set of bacteria cells with bacteriophages with 32P labeled DNA (shown here as blue). The bacteria and the bacteriophages were mixed in a tube, after some time they shook the tube to release the bacteriophages from bacterial surface of course first giving the bacteriophage enough time to inject their genes in the bacteria. Then they centrifuged the bacterial cells and looked for the presence of radioactivity – in supernatant and in the pellet. What did they find? The found that they could recover radioactivity inside bacterial cells only when they used 32P labeled bacteriophages. This indicated that it was the DNA that was injected inside bacteria and not protein! Yay! Definitely check out the animation for this experiment.
Watson and Crick’s semiconservative model of replication predicts that when a double helix replicates, each daughter molecule will have one old strand (derived or “conserved” from the parent molecule) and one newly made strand. Competing models were the conservative model (the two parent strands rejoin) and the dispersive model (each strand is a mix of old and new)
Bacterial cultures grown in regular 14N or 15N are the controls. After several rounds of growing bacteria in 15N, they were transferred to medium containing 14N. After they were grown for about 20 min (one round of replication), bacteria were removed, DNA extracted from these bacteria and subjected to centrifugation with CsCl. They combined the DNA with CsCl – and spun at high speeds in an ultracentrifuge. This resulted in forming a gradient of CsCl concentrations. The density of CsCl was highest at the bottom of the tube and lowest at the top. DNA in this solution formed a band at the point in the tube where the density of CsCl corresponded with its density.
So three bands distribution are possible – high density (bottom of tube), intermediate density (middle of the tube), low density (top of the tube). After the first round of replication they noted the distribution of bands. Based on what they observed after one round of replication (one band with intermediate density) they could eliminate conservative model of replication, as you would expect a high density band as well for this model. After the second round of replication they were able to eliminate the dispersive model. So the correct model of replication is semiconservative!
Structures of purines and pyrimidines were connected by hydrogen bonds.
Based on Chargaff’s data, X ray diffraction pattern of DNA by Rosalind Franklin, Watson and Crick came up with the double helix model of DNA. Remember this from chapter 3 – note base complementarity. The way this structure would satisfy the X ray image was if the two strands were antiparallel.
The amount of Guanine equals the amount of Cytosine. The amount of Adenine equals the amount of Thymine.
Subtopic
Central Dogma of Molecular Biology
DNA, RNA, PROTEIN
DNA
Structure: Double-stranded helix
Function: stores genetic information
Transcription
Initiation
RNA Polymerase: enzyme that initiates transcription
Promoter Region: specific DNA sequence signaling the start of transcription
Elongation
RNA Synthesis: RNA polymerase reads the DNA template and synthesizes complementary RNA
Template strand: the DNA strand used as a template for RNA synthesis
Termination
Terminator sequence: DNA sequence signaling the end of transcription
Release of RNA: RNA polymerase and the newly formed RNA molecule detach from the DNA template
RNA produced
mRNA: carries the genetic information from DNA to the ribosome
tRNA: transfer amino acids to the ribosome during translation
Translation
Site: ribosomes, involves tRNA bringing amino acids to the ribosome
Initiation
Start codon: AUG on mRNA signals the beginning of translation
ribosome binding: small ribosomal subunit binds to mRNA at the start codon
Elongation
Amino acid addition: tRNA molecules bring amino acids to the ribosome
Codon Recognition
Termination
Stop codon signals the end of translation
release factor: binds to the stop codon, causing the release of the polypeptide chain
Protein Synthesis
codons: three-nucleotide sequences on mRNA
Genetic code: the correspondence between codons and specific amino acids
disassembly: ribosome dissociates into its subunits
Peptide bond formation: Ribosome catalyzes the formation of peptide bonds between amino acids
Translocation: ribosome moves along mRNA, reading codons and synthesizing the growing polypeptide chain
tRNA binding: initiator tRNA carrying methionine binds to the start codon
rRNA: major component of ribosomes
Complementary base pairing: A-U, G-C in RNA synthesis
Transcription factors: proteins that assist in the binding of RNA polymerase to the promoter
Processes: Replication
DNA Replication
Watson and Crick noted that the specific base pairing suggested a possible copying mechanism for genetic material where each strand serves as a parent strand with information to make the other strand. Based on this as it retains the parent and uses this information to form a new (daughter) strand this was coined as the semi conservative model. This model – semiconservative replication was proposed, But is this the correct model? There were competing models proposed as well.
There is a sequence of nucleotides in DNA called ORI for origin of replication. The goal is to separate the two strands of DNA at ORI sequences and form a bubble as shown here. This is called a replication bubble and there are two forks at each end of the bubble. Now each separated strand here has to be used as the parent strand to form complementary daughter strand starting at the ORI sequence, as suggested by the semiconservative model.
For the long DNA molecules in eukaryotes, multiple replication bubbles form and eventually fuse, speeding up the copying of DNA, while as Prokaryotic DNA is circular there is only one ORI sequence.
An enzyme Helicase separates the two strands to form the replication bubble. SSB or single stranded proteins keep the DNA single stranded. Another enzyme Topoisomerase helps relieve any strain caused by unwinding of the DNA.
Primase – makes RNA primers complementary to the DNA parent strand sequence. Now the DNA polymerase III will add nucleotides only to the 3’ end. Note the arrow.
DNA polymerase III. This enzyme will add nucleotides only to the 3’ end.
DNA polymerase III is the enzyme that makes daughter DNA strands, but first it needs a short (20 nucleotides or so) strand to add new nucleotides to. This is called a primer. In replication this is an RNA primer. The DNA polymerases adds new nucleotides complementary to parent DNA, to the primer in ONE direction only – 5’ to 3’, so always adding nucleotides to the 3’end of a previous nucleotide. Another protein called Sliding Clamp works with DNA polymerase III. It helps keep DNA pol III on the parent strand so it doesn’t fall off during replication.
So to form a strand we have to connect nucleotides through phosphodiester bonds using dehydration/condensation reactions! Yay for chapter 3! We need enzymes for this. The enzyme here is a DNA polymerase. In bacteria it is DNA polymerase III. But for a DNA polymerase to add nucleotides it has two constraints: It adds nucleotides ONLY to the 3’ end of an existing nucleotide. Which means it adds nucleotides in 5’ to 3’ directions and needs something to add nucleotides to – we call this a primer. Typically in cells the primer for replication is a short sequence of RNA and the enzyme that makes an RNA primer is called a Primase.
Along one template strand of DNA, the DNA polymerase synthesizes a leading strand continuously, moving toward the replication fork. Only one primer is required to synthesize the leading strand. Again keep in mind the strand is made in one direction 5’ to 3’
In forming the lagging strand, multiple RNA primers have to be laid down and then extended by DNA polymerase III to form short Okazaki fragments. So how are these RNA primers removed? Another enzyme, DNA polymerase I removes the RNA and replaces it with DNA nucleotides. Another enzyme called ligase seals any gaps by connecting nucleotides by phosphodiester linkages.
. Each fork moves in the opposite direction hence replication is called bidirectional. Blue strands of DNA are the daughter strands. So what are the red lines? That’s the RNA primer. The arrows on the light blue strands tell us the direction in which new nucleotides are added. So in this case when the nucleotides are added continuously in the direction of the fork that strand is called the leading strand. Now as DNA is antiparallel, the opposite strand will be made in the opposite direction, right? Yes, but the opposite strand is formed in short segments called Okazaki fragments, and is called the lagging strand. In bacteria, DNA polymerase III forms these new strands through condensation synthesis.
Subtopic
Topic principal
mitochondria
organelle where
cellular respiration occurs and
most ATP is generated
Lysosome
digestive
organelle where
macromolecules are
hydrolyzed
Phagocytosis
1.Lysosome contains active hydrolytic enzymes
2. Lysosomes fuses with food vacuole
3. Hydrolytic enzymes digest food particles
Autophagy
1. Lysosomes fuses with vesile contaning damaged organelles
2. Hydrolytic enzymes digest organelle components
Plasma mebrane
membrane enclosing the cell
Subtopic
Cell's Signaling
long distance signaling
physical contact
occurs in neighboring cells that are in physical contact.
the signaling molecule is not free, it is bound to the membrane cell
interacts with receptor on the membrane of adjacent cell
Local signaling
Realeases a signal ( requires the target cell to have sometging to receive signal
Paracrine signaling
Synaptic signaling
Receptor
Present in target cell that receives the signal molecule
Intracellular receptors: In cytoplasm & nucleus
Membrane receptors : hydrophilic- first messenger & requires a second messenger
G-protein linked receptor
when the signaling molecule binds to the extracellular side of the receptor, the receptor is activated and changes shape. Its cytoplasmic side then binds and activates a G protein. The activated G protein carries a GTP molecule.
The activated G protein leaves the receptor, diffuses along the membrane, and binds to an enzyme, altering the enzyme's shape and activity. Once activated the enzyme can trigger the next step leading to a cellular response. Binding of signaling molecules is reversible. The activating change in the GPCR, as well as the changes in the G-protein and enzyme, are only temporary; these molecules soon become available for reuse.
tyrosine kinase receptor
Ion channel receptors
Golgi Apparatus
Organelle active
in synthesis, modification, sorting,
and secretion of cell products
Cytoskeleton
reinforces cell's shape; functions in cell movement
Microfilaments
Intermediate filaments
Microtubes
Microvilli:
projections that
increase the cell’s
surface area
Ribosomes
complexes that
make proteins; free in
cytosol or bound to
rough ER or nuclear
envelope
Microfilaments
Intermediate filaments
Microtubules
Endoplasmic reticulum
network
of membranous sacs and tubes; active in
membrane synthesis and other synthetic
and metabolic processes; has rough
(ribosome-studded) and smooth regions
Rough ER
Smooth ER
Chloroplast
photosynthetic
organelle; converts energy of
sunlight to chemical energy
stored in sugar molecules
Peroxisome
organelle with
various specialized metabolic
functions; produces hydrogen
peroxide as a by-product and
then converts it to water
Cell Wall
outer layer that maintains
cell’s shape and protects cell from
mechanical damage; made of cellulose,
other polysaccharides, and protein
PROKARYOTES
Plasma Membrane and Nucleoid
Bacterial chromosome, nucleoid, is in the cytoplasm because there is no nucleus. The presence of DNA that is separate from the main bacterial chromosome is plasmid (made of proteins and phospholipids).
Cell Wall
This is the structure present outside of the plasma membrane. Functions include: provides bacteria with support, protects the cell, and maintains shape.
Capsules and Slime Layer
The cell wall of many bacteria is surrounded by a sticky layer made of polysaccharide or protein. The capsule is dense and well defined or a slime layer if diffuse. Both of these outer layers help bacteria to stick (adhere) to a substance or other individuals in a colony. These layers also protect against dehydration and some capsules protect bacteria from attacks by host's immune system.
Flagellum
Flagella of bacteria , archaea and
eukarya differ in their composition
and mechanics of movement. Flagellum contains a hook and basal body. It rotates 360 degrees to propel the cell.
Fimbriae and Pili
Some bacteria contain on their surface short hair like projections called fimbriae. Some bacteria also contain long projections called pili which are involved in forming a channel between two bacterial cells to transfer DNA from one cell to another.
Ribosomes
Complexes that synthesize protein.
Made up of two RNA complexes.
Cellular Respiration
steps
Glycolysis
Glucose
Glucose 6-phosphate
Fructose 6-phosphate
Fructose 1,6-biphosphate
Glyceraldehyde 3-phosphate
1,3-bisphosphoglycerate
3-phosphoglycerate
2-phosphoglycerate
phosphoenolpyruvate
Pyruvate
Pyruvate Oxidation
Pyruvate
Acetyl CoA
Pyruvate
Acetyl CoA
Citric Acid Cycle / Kreb's Cycle
Acetyl CoA
Citrate
5-carbon molecule
4-carbon molecule
GDP, Pi to GTP
FAD to FADH2
NADH to NAD+
Oxaloacetate
Oxidative Phosphorylation
Dihydroxyacetone phosphate
DNA Replication
Subtopic
Cell
Plasma mebrane
Subtopic
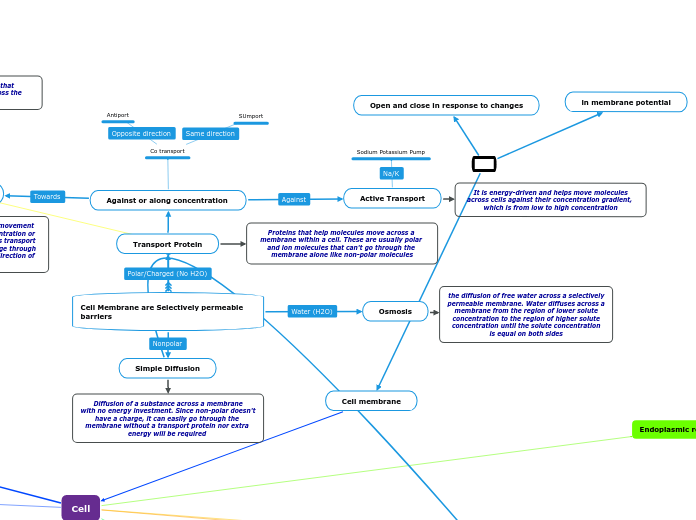
Cell Membrane are Selectively permeable barriers
Transport Protein
Against or along concentration
Co transport
Antiport
SUmport
Facilitated Transport
Carrier Protein
It undergo a subtle change I shape that
translocate the solute binding site across the membrane
A type of diffusion which means that movement
of molecules is from high to low concentration or down concentration gradient. It speeds transport of a solute by providing efficient passage through the membrane but does not alter the direction of transport
Channels
Gated Channels
Voltage-Gated
Ligand-Gated
Stretch-Gated
Provide corridors or channels that allow
a specific molecule or ion to cross the membrane
Non-gated Channels
Allows molecules to pass by under
any circumstances
Active Transport
Sodium Potassium Pump
It is energy-driven and helps move molecules
across cells against their concentration gradient, which is from low to high concentration
Proteins that help molecules move across a membrane within a cell. These are usually polar and ion molecules that can't go through the membrane alone like non-polar molecules
Osmosis
the diffusion of free water across a selectively
permeable membrane. Water diffuses across a
membrane from the region of lower solute
concentration to the region of higher solute
concentration until the solute concentration
is equal on both sides
Simple Diffusion
Diffusion of a substance across a membrane
with no energy investment. Since non-polar doesn't have a charge, it can easily go through the membrane without a transport protein nor extra energy will be required
Electron transport chain
Cell Regulation
Positive Regulation
Active repressor binds
No transcription
Active repressor doesn't bind
to operator
Operon "on"
Operon OFF
Operon "on"/
transcription
Operon ON
Operon OFF
Eikaryotic ONLY
cant bind so no transcription
Transcription factors bind
before RNA Polymerase II
RNA Polymerase II binds
Additional Transcription factors
bind along with RNA polymerase II
Transcription/DNA unwinds
Negative regulation
Activator is Bonded to
operator
Activator is not bonded
to operator
transcription
Cell Wall
outer layer that maintains
cell’s shape and protects cell from
mechanical damage; made of cellulose,
other polysaccharides, and protein
V
Biomolecules
Carbohydrates
Polysaccharids
Starch
Amylopectin
Amylopectin: A branched polysaccharide that is stored in starch, it stores glucose for later use for energy source. The branching helps glucose units to be synthesized and stored.
Amylose
Amylose: Amylose is a unbranched polysaccharide chain. It helps plant store energy and break up starch molecules in maltose and maltotriose.
Cellulose
Cellulose: It is an unbranched polysaccharide that is used for structure in plants. Being unbranched makes it able to form a long and straight chain which allows rigidity in the plant cell and maintain its shape.
Glycogen
Glycogen: A long polymer chains of glucose that are bonded with an alpha acetal linkage (1-4). It has branching at after every 4-8 residues and is bonded by a 1-6 glycosidic bond. Glycogen is used as a storage that allows fast energy release when it is needed. The branchings makes it possible for rapid hydrolysis which allows more respiration during exercises.
Monosaccharides
Alpha Glucose
Alpha glucose: Hydroxyl group is facing down from the main structure. This makes it more reactive to enzymes and less stable. Glycogen and starch are made up of alpha glucose.
Beta Glucose
Beta glucose: Hydroxyl group is facing above the main structure. Cellulose is made up of beta glucose.
Fructose
Fructose: It has the same chemical formula as glucose just different structure. It is found in fruits, vegetable, honey, and more. Fructose helps provide energy when glucose is not sufficient. Compared to glucose, Fructose is a five-membered ring.
Galactose
Galactose: Another isomer of glucose, which means same formula but different structure. The main difference is the hydroxyl arrangement at carbon four. Compared to glucose, galactose is not as sweet as glucose and less soluble in water.
Lipids
Saturated Fats
Saturated Fats: Consist of a glycerol and three fatty acids, or is called a triglyceride. The fatty acids in saturated fats are all single bonds between carbon and hydrogen. This makes the structure straight and packed. Which means in real life, food with more saturated fat in room temperature will become solid.
Unsaturated Fats
Unsaturated Fats: The fatty acids in unsaturated fats have some double bonds between carbon and hydrogen which means the acids aren’t as packed and more spread out compared to saturated fats. In room temperature, foods with unsaturated fats will stay in liquid form.
Trans Fat
Trans Fat: Around the double bond of carbon in the fatty acid, there will be two hydrogens on the opposite sides. This makes it straighter than cis fats. Since it is straighter, it will be more solid than cis fats which means it can also become solid at room temperature. Trans fat is usually a result of trying to transform a liquid oil to solid. This makes it harder to digest and break down and can cause a increase level in LDL (low-density lipoprotein) or bad cholesterol.
Cis Fat
Cis Fat: Hydrogens around the double bond will be on the same side which means the acid would be more bent, making it less solid and packed. This makes it liquid at room temperature and easier to digest. Making it the healthier option to consume.
Nucleic Acid
(RNA) Ribonucleic Acid
Ribonucleic Acid (RNA): RNA is made up of one strand or single helix. Its main function is to create proteins and carry genetic information that is translated by ribosomes. Because it is a single helix, it is less stable making it not as reliable compared to DNA whenever it comes to the stability in genetics. There are three main types of RNA: mRNA, rRNA, and tRNA.
DNA (Deoxyribonucleic Acid)
Deoxyribonucleic Acid (DNA): DNA is made up of two strands that wrap or wind up around each other, which is also called a double helix. These two strands are connected together with hydrogen bonds. It also replicates and store genetic informations. The reason people use DNA more than RNA when it comes to genetics is because DNA is more stable which makes it safer when it comes to keeping genetic information.
Proteins
Tertiary
Non-Polar
Non-polar: The end of the R group is a non-polar component like H, CH, or a hexagon ring. If proteins are made up of non-polar R groups, it would mostly be found tucked in when reacting with water because of being hydrophobic reaction.
Polar
Polar: The end of the R group is a polar component like OH, SH, and NH2. They would usually be found on the outside when reacting with water because they are hydrophilic. They would interact with each other through dipole-dipole, covalent, or hydrogen bonds.
Charged
Charged: The R group contains charges either an acidic (negative) or basic (positive). They would react with opposite charges through ionic bonds.
Tertiary: the third level of protein level. In tertiary level, R chains are involved in bondings. Non-polar would interact through hydrophobic interactions. Polar would interact with dipole-dipole, disulfide, covalent, hydrogen bonds, or hydrophilic interactions. Charged groups will interact with ionic interactions. Tertiary structure also determines how hormone regulates receptor activations. In this phase, polypeptide chains become functional. It becomes three-dimensional and allows it to interact with other molecules.
Quaternary
Quaternary: This level occurs with tertiary structures interact with each other through their R chains. The way they interact also depends on if they are non-polar, polar, or charged. At this level, it would allow proteins to have multiple functions in gene expression, macromolecular transport, and chemical reaction regulation. However, not all protein have quaternary structures.
Secondary
Beta Pleated Sheets
Beta Pleated Sheets: this cannot be a single polypeptide because it needs one or more strands. It is crucial for lipid metabolism and the structure of proteins that binds with fatty acids. They also bond together through hydrogen bonds but instead of forming spirals, they are parallel to each other.
Alpha Helices
Alpha Helices: this can be a single chain polypeptide. Alpha helice occurs when the polypeptide winds up and forms a spring-like structure. The structure is held together through hydrogen bonds. It is most used secondary structure and is used in the protein-DNA interface interaction. It also determines the global structure of proteins and their functions.
Secondary: This is the secondary level folding. Here there will be two types of structure:
Primary
Primary: Primary is the first level of protein fold which mean it is just a long string of amino acids bonded together at the main chain by peptide bonds.
Open and close in response to changes
in membrane potential
Fimbriae are shorter than
pili and more numerous.
The cell wall is made up of
peptidoglycan in bacteria.