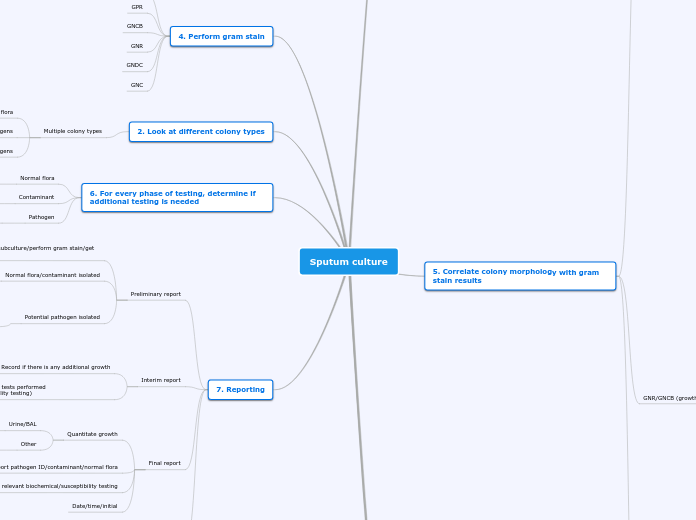
Sputum culture
3. Examine colony morphology
White/gray, smooth, raised, beta or gamma, opaque
Think possible GPC in clusters or chains
Gray, smooth, pinpoint, flat/raised, alpha, translucent
Think possible GPC in chains
Gray/tan/bluish-gray/white, small, translucent, raised, convex, smooth, hockey puck
Think possible GNDC
Wide variety of colony morphologies
GNR/GNCB
5. Correlate colony morphology with gram stain results
GPC
Perform catalase
+
Perform coagulase
+
Staph aureus
-
Perform bacitracin or modified oxidase
Bacitracin S/modified oxidase +
Micrococcus species
Baccitracin R/modified oxidase -
(Other) coag neg staph
(Urine) novobiocin
S
Coag neg staph
R
Probable staph saprophyticus
-
Possible streptococci
Beta
Perform PYR
+
Perform bacitracin
S
Group A strep
-
Perform hippurate hydrolysis
+
Group B strep
-
Perform bile esculin
+
Perform 6.5% NaCl
+
Enterococcus species
-
Group D strep
-
Beta strep, not A, B, or D, or Enterococcus species
Alpha
Perform optochin
S
Strep pneumoniae
R
Perform bile esculin
+
Perform 6.5% NaCl
+
Enterococcus species
-
Group D strep
-
Strep viridans
Gamma
Perform bile esculin
+
Perform 6.5% NaCl
+
Enterococcus species
-
Group D strep
-
Non-hemolytic strep
GNR/GNCB (growth on BAP, CHOC, & MAC)
Growth on MAC
+
Perform oxidase
+
G
Glucose fermenter
Correlate with key biochemical reactions
Glucose oxidizer
ADH
+
Growth @ 42 C
+
Pseudo aeruginosa (if green/red/brown pigment, LDC -, ODC - , nitrate +)
-
Additional results needed for definitive ID
-
LDC
+
Perform polymyxim B susceptibility
S
Additional test results needed for definitive ID
R
Burk cepacia complex (if ODC variable, pale yellow colonies)
-
Probable Elizabeth meningoseptica (if ODC -, indole +, yellow pigment)
Glucose asaccharolytic
Additional test results needed for definitive ID
-
(Sterile body source, LF) perform GNR ID panel
(Non-sterile body source, LF) observe lactose fermentation
(Non-GI site) perform spot indole
+
Beta
+
Esch coli
-
Perform PYR
+
Perform GNR ID panel
-
Esch coli
-
Perform GNR ID panel
(GI site) perform GNR ID panel
NLF
Observe swarming on BAP
+
Perform spot indole
+
Prot vulgaris
-
Perform GNR ID panel
Ampicillin S
Prot mirabilis
Ampicillin R
Perform GNR ID panel
ODC +
Prot mirabilis
ODC -
Prot penneri
-
Perform GNR ID panel
Glucose fermenter
Correlate with key biochemical reactions
Glucose oxidizer
LDC
+
Streno maltophilia (if ADH/ODC -, strong maltose oxidizer, pale yellow/lavendar colonies on BAP)
-
Probable Acinetobacter species (if ADH/ODC -, plump GNCB, colorless/pink colonies on MAC)
Glucose asaccharolytic
-
Growth on BAP
+
-
GNR/GNCB (growth with only fastidious organisms)
Perform catalase
+
Perform oxidase
+
Perform XV tests on horse BAP
XV growth
Beta
+
Haem haemolyticus
-
Haem influenzae
X growth
Haem species
V & XV growth
Beta
+
Haem parahaemolyticus
-
Haem parainfluenzae
-
Consider other organisms/repeat testing
-
Perform gram stain
GNCB with school of fish/railroad tracks
+
Suspect Haem ducreyi
Perform XV tests on horse BAP
X & XV growth
Haem ducreyi
-
Consider other organisms
1. First & foremost = always consider source type
May change battery of tests performed
7. Use abbreviated ID if possible
4. Perform gram stain
GPC
GPR
GNCB
GNR
GNDC
GNC
2. Look at different colony types
Multiple colony types
Normal flora
Pathogens
Mixture of normal flora & pathogens
6. For every phase of testing, determine if additional testing is needed
Normal flora
Discontinue testing
Contaminant
Discontinue testing
Pathogen
Continue testing
7. Reporting
Preliminary report
Unable to isolate colonies/subculture/perform gram stain/get colonies to grow
Report as culture ID pending
Normal flora/contaminant isolated
Report as normal flora/possible contaminant & quantitate
Potential pathogen isolated
Report relevant info (i.e. LF GNR) or use abbreviated ID & quantitate
Pathogen ID unknown or pathogen antibiotic susceptibilities unknown = report as antibiotic susceptibility/ID pending
Pathogen ID known or antibiotic susceptibilities known = report as is & do not perform susceptibility testing (i.e. beta hemolytic strep is known to be susceptible to penicillin)
Interim report
Record if there is any additional growth
Run & record any more tests performed (biochemical/susceptibility testing)
Final report
Quantitate growth
Urine/BAL
Quantitate numerically (i.e. >100,000 CFU/ml)
Other
Quantitate qualitatively (i.e. many, few, etc.)
Report pathogen ID/contaminant/normal flora
Note relevant biochemical/susceptibility testing
Date/time/initial
ALWAYS date/time/initial reports, beginning of overnight tests, susceptibility testing, etc.