por Melody Tran 12 meses atrás
147
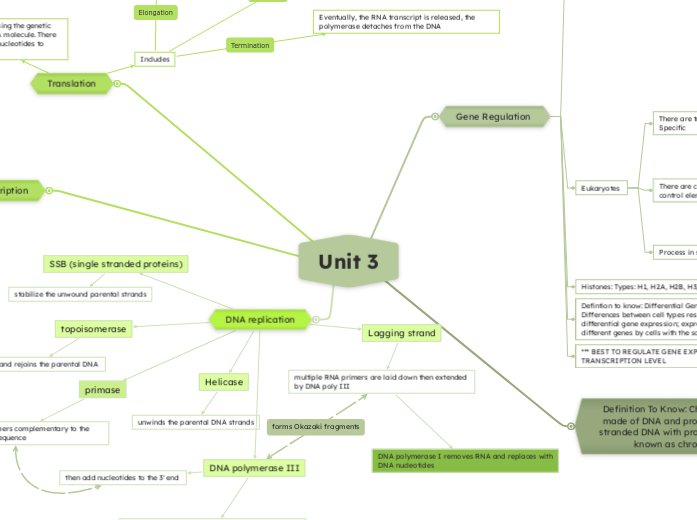
Unit 3
por Melody Tran 12 meses atrás
147

Mais informações
DNA polymerase I removes RNA and replaces with DNA nucleotides
The transcription of a protein-coding eukaryotic gene
A sequence within a primary transcript that remains in the RNA after RNA processing; also refers to the region of DNA from which this sequence was transcribed.
A noncoding, intervening sequence within a primary transcript that is removed from the transcript during RNA processing; also refers to the region of DNA from which this sequence was transcribed.
3 Active Sites
One of a ribosome’s three binding sites for tRNA during translation. The E site is the place where discharged tRNAs leave the ribosome. (E stands for exit.)
One of a ribosome’s three binding sites for tRNA during translation. The P site holds the tRNA carrying the growing polypeptide chain. (P stands for peptidyl tRNA.)
Step 1: Activator proteins bind to distal control elements
Step 2: A DNA-bending protein brings activators closer to promoter, there are general transcription factors, mediator proteins, and RNA polymerase II
Step 3: Activators bind to certain mediator proteins and general transcription factors
Distal control elements: There are enhancers and bind to specific transcription factors (activators and repressors)
Location: Sequences in DNA upstream or downstream of gene; possibly close or far away from gene
Proximal control elements: sequences in DNA close to promoter as well as bind general transcription factors
Specific: Activators and repressors involved to either increase levels of transcription or decrease
General: background/basal level of transcription
CAP is activated by cAMP; cAMP binds to CAP and helps RNAP bind to promoter to facilitate transcription = operon is on!
**If there is no cAMP because of the presence of glucose that blocks Adenylyl Cyclase, CAP cannot be activated and if CAP is not active, it cannot help RNAP bind to the promoter = operon is OFF!
When no lactose is present, Lac I makes a Lac repressor proteins that binds to the operator sequence and prevents expression of the lac operon structural genes
If there is also no glucose and no lactose present, the operon is ON!
When lactose is present, the gene expression will be turned on because Lac repressor protein binds to lactose; as a result, RNAp can bind to promoter
Organization of lac operon: DNA, promoter, regulatory gene, promoter for structural genes, operator, Z,Y,A genes
Structural genes for Lac Operon: LacZ, LacY, LacA
LacA: Trans-acetylase
LacY: Permease
LacZ: B-galactosidase
Negative Regulation: When repressor is bound to the operator, no transcription occurs; when no repressor is bound, transcription occurs
Positive Regulation: When activator is bound to the operator, transcription occurs; when no activator is present, no transcription will occur