Gene expression in the Three-Spined Stickleback (Gasterosteus aculeatus) of Marine and Freshwater Ecotypes

Gene expression
In total, 2,982 differentially expressed genes between marine and freshwater populations were discovered

The RNA concentration for each sample was determined using a BioAnalyzer 2100 (RNA 6000 Nano Kit) (Agilent, USA)

To identify the genes which are differentially expressed in the samples of threespined stickleback, the Illumina nucleotide reads were mapped on the G. aculeatus reference genome from the Ensembl database.

Differential expression was established using the edgeR package, which calculates the variance of the expression index for each gene.
The total number of reads of 75 nucleotides in length was 85438630 and 110690898 in the libraries from the marine and freshwater samples, respectively.

the Three-Spined Stickleback
A marine population of three-spined stickleback usually uses freshwater streams and lakes for spawning
Samples of marine three-spined stickleback (hereinafter referred to as “M”) were collected from:
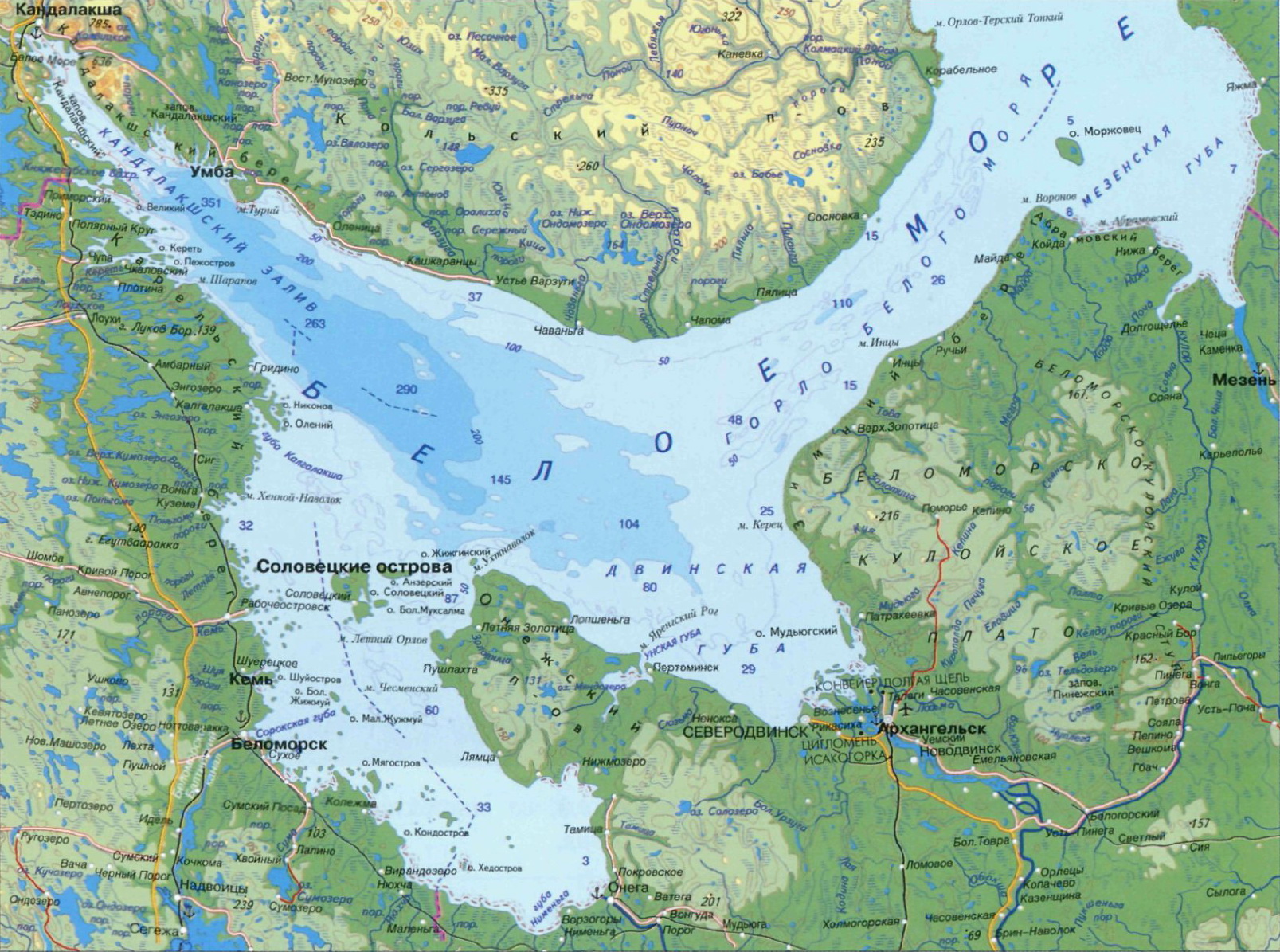
The White Sea, near the N.A. Pertsov White Sea Biological Station of Moscow State University.

Freshwater samples were collected from the Machinnoye Lake, not far from the village of Tchkalovsky, Loukhskiy district, Republic of Karelia.