STRUCTURE MAKES FUNCTION
Macromolecules
Nucleus Acids
DNA
RNA
Nucleotide
Lipids
Phospholipids
Phospholipid bilayer
Trigycerol
Saturated fats
Energy source (butter)
Unsaturated fats
Important Energy source (oils)
Steroids
Cholesterol
Testosterone (hormones)
Carbohydrates
Monosaccharide
Fructose
Glucose
Disaccharide
Lactose
Sucrose
Polysaccharide
Storage
Starch
Glycogen
Structural
Cellulose
Starch
Proteins
Enzymes
Structural
Storage
Transport
Hormones
Receptor
Motor
Defensive
Eukarytotes

Chemistry of life
Genes
Regulation
Level
transcription
RNA Processing
Transport to cytoplasm
Translation
Protein Processing
Expression
Prokaryotes
Energy
Metabolism
Free Energy
Equilibrium
ΔG = 0 , Equilibrium
Max stability = Equilibrium
Free energy decreases closer to equilibrium & Vice Versa.
System never moves away from Equilibrium Spontaneously
Equilibrium= Dead System
ΔG
Universe = System + Surroundings
When temp. and pressure are uniform
ΔG = ΔH -TΔS
ΔG> 0, non-spontaneous, endergonic
ΔG< 0 , spontaneous, exergonic
ΔG = ΔG final - ΔG initial
Images


Subtopic
Energy Transformation
Metabolic Pathways
Catabolic Pathway
Anabolic Pathway
Energy
Kinetic Energy
Thermal Energy
Heat: transfer of thermal energy
Chemical Energy
Thermodynamics
Surrounding vs. system
1st Law: Principle of Conservation of Energy
2nd Law: Energy transfer increases entropy of Universe
Entropy and Enthalpy
Large amount of energy transferred is lost as heat
Coupling
Work
Chemical Work
Transport Work
Mechanical Work
Phosphorylation
Phosphorylated intermediate: key to coupling
Coupling occurs by endergonic and exergonic reactions fueling each other.
Enzymes
Activation Energy
Energy required to get to the "top of the hill"
Transition state
Has enough energy to break bonds
How they work
Catalyst
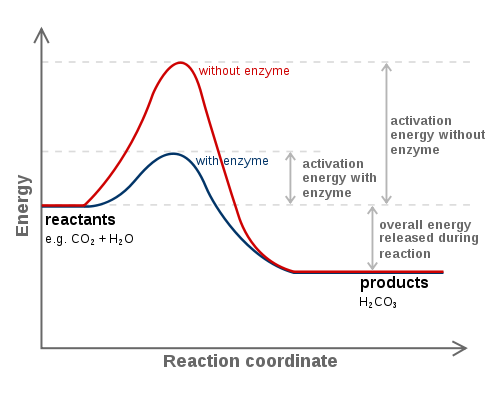
They lower Ea
Enzyme -substrate complex
Converts substrate to products
Induced fit via malleable equilibrium
Optimal conditions
pH
Temperature
Cofactors
Non protein helpers
Competitive inhibitors
Bind to active site
Non competitive inhibitors
Bind somewhere other than active site
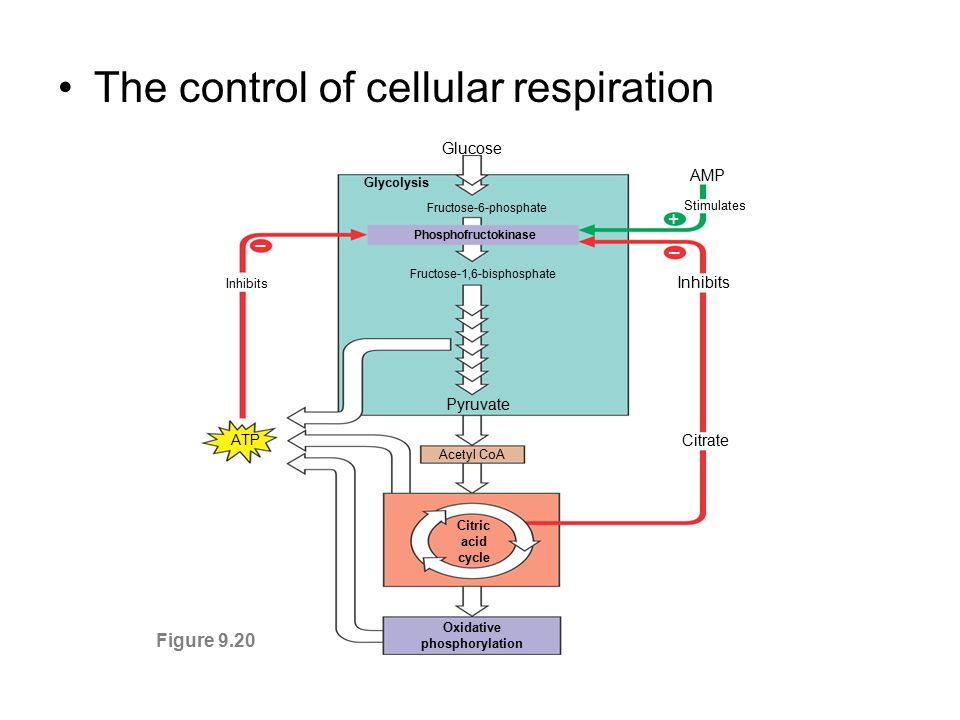
Enzyme Regulation
Allosteric Regulation
Not bound on active site
Oscillating subunits
Activators
stabilize subunits
Inhibitors
stabilize subunits
Enzymes and substrates are compartmentalized
within the cell
Cooperativity
allosteric activation
substrate binds to active sites
amplifies enzyme response
affects other binding sites
Cellular Respiration
Catabolic Pathway, redox
OIL (Reducing agent)
RIG (oxidizing agent)
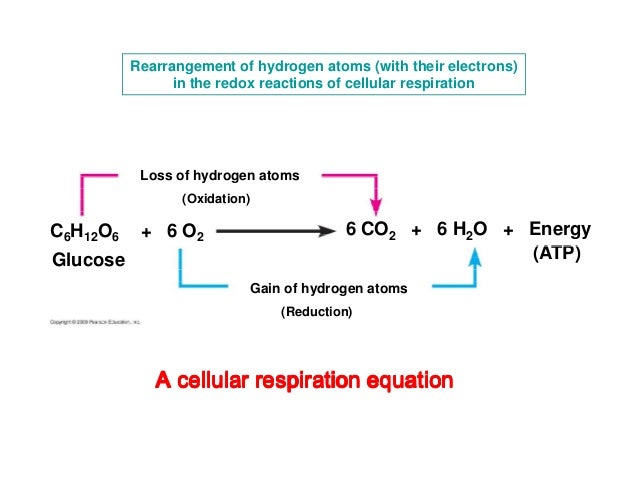
NAD+ oxidized state
NADH reduced state
Substrate-level phosphorylation
Phosphate for ADP comes from substrate
rather that an inorganic phosphate
Substrate-level phosphorylation
Powered by redox reactions
Glycolysis
Energy Investment Phase
Glycolysis occurs whether or not O2 is present
Step 1
Hexokinase transfers phosphate from ATP to Glucose
Step 3
Phosphofructokinase transfers a phosphate from ATP to opposite end of the sugar (this is the second ATP)
Step 5
Two three carbon sugars are left
(Glyceraldehyde 3-phosphate and
Dihydroxyacetone Phosphate)
G3P and DHAP which convert between each other
Energy Payoff Phase
4 ATP formed, 2 NADH, 2 H+, 2 pyruvate, 2 H2O
Net
Glucose --> 2 Pyruvate + 2 H2O
2 ATP, 2 NADH, + 2 H+
Occurs in the cytosol
Pyruvate oxidation
Occurs in the mitochondrial matrix
Step 1
Pyruvate dehydrogenase complex removes the CO2 from the pyruvate.
Methionine first amino
acid placed in protein
Step 2
Remaining fragment is oxidized forming acetate and releasing electrons forming NADH.
Step 3
Acetyl CoA is formed via coenzyme A
Citric acid cycle
Two reduced carbons enter the cycle with Acetyl CoA, and two oxidized carbons leave in the form of CO2
Step 1
Acetyl CoA becomes citrate
Step 3
Isocitrate is oxidized, reducing NAD+ to NADH and losing a CO2 molecule. Isocitrate becomes a-ketoglutarate
Step 4
Another CO2 is lost, NAD+ becomes NADH. a-ketoglutrate becomes Succinyl CoA
Total yield
One glucose--> 6 NADH, 2FADH2, 2 ATP
Oxidative phosphorylation
Electron Transport Chain
When a complex receives en electron, it is reduced, then oxidized when it is passed onto the next complex
The electrons travel from complex I and II to complex Q, then to Complex 3 followed by complex 4.
Total: 2 e-, 2H+, 1/2 O2
Occurs in the inner mitochondrial membrane
Electron carriers
NAD+
FADH2
Cytochromes
Chemiosmosis
ATP Synthase
makes ATP from ADP and inorganic phosphate
H+ concentration gradient on the outside of the inner membrane couples the redox reactions of the ETC
26 or 28 ATP produced
Anaerobic respiration
Fermentation
Alcohol fermentation
Pyruvate is converted to ethanol
Lactic acid fermentation
pyruvate is reduced by NADH to form lactate
Photosynthesis
Photosynthesis makes food
autotrophic
Makes its own food
Choloroplasts
Thylakoid
Where light reactions occur
split water
release O2
Produce ATP and form NADHP
grana
Stroma
Calvin cycle
CO2--> sugar
Uses ATP for energy
Uses NADPH for reducing power
6CO2 + 12H2O + Light Energy --> C6H12O6 + 6O2 + 6H2O
Photosynthesis is a redox
H2O is oxidized
CO2 is reduced
Light reactions
Wavelengths
Visible light wavelengths drive photosynthesis
Photon
Pigment absorbs
Photosytems
PS I
P700 molecules
PS II
P680 molecules
Both contain a reaction-center complex and a light harvesting complex
Electron flow
Linear
Uses both PS's and produces, NADPH, ATP, AND O2
Cyclic
only one PS is used, produces only ATP
Calvin Cycle
Uses ATP and NADPH to reduce CO2 and sugar
One molecule of G3P exits per three CO2
Alt. mechanisms for Carbon Fixation
Photorespiration
O2 subs for CO2 in rubisco sites
C3 plants
Close stomata conserving water
C4 plants
Incorporate CO2 in mesophyll cells
then exported to bundle-sheath cells
CAM plants
open their stomata at night
Lac operon
Trp operon
Prokaryotes
Formylmethionine first
amino acid in protein
Switch: Operator
ON
Positive
Activator
Increased RNA transcription
OFF
Negative
Repressor
Basal level
Structural
Lac Z
Beta Galactosidase
Lac Y
Permease
Lac A
Beta Galactosidase acetylase
Trp E
Trp D
Trp C
Trp B
Subtopic
Trp A
Regulatory
Lac I
Trp R
Constituive Expression
Lactose present
Glucose scarce
CAMP levels high
When all conditions met
Lactose absent
repressor unbound
When conditions are met
Glucose high
CAMP low
Repressor bound
Polypeptide subunits
Protein
Enzymatic
Transport
Cell cell recognition
signal transduction
Inter cellular joining
ECM/ cytoskeleton
Tryp. absent
Repressor not bound
Tryp. present
Repressor bound
Eukaryotes
Histone packaging
Allows coiling of DNA
DNA organized neatly in nucleus
Methylation
Acetlyation
Specific transcription factors
General transcription factors
control elements
Distal
Proximal
Closes chromatin
Opens up chromatin
Amino acid tails exposed
Accessible for modification
Corepressor
Are examples of
Functions include:
Glycolipids
Glycoproteins
Carbohydrate markers
Uneven distribution of carbs.
Carrier
Two conformations
Channel
Na/K pump
Affinity for
K
Na
OR
Maintain concentrations- substance/solute
Active transport
Simple diffusion
facilitiated diffusion
Tonicity
Plant
Animals
Isotonic
Normal
Hypotonic
Shrivel
Hypertonic
Lyse
Relative to cell
Isotonic
Flaccid
Hypotonic
Turgid
Ideal conditions
Hypertonic
Plasmolyzed
Electrogenic pump
Proton Pump
Work possible
Energy
Affects Membrane
Phospholipid membranes
Lipids
Carbs
Main Ingredients
Across Gradient
Active Transport
Requires Energy
Cholesterol
Affects fluidity
regulates membrane fluidity
Amphipathic
Hydrophobic exterior/ hydrophilic interior
Large
Polar
Charged
Hydrophilic
Allows passage of
Gradient
Cotransporters
Powers
ATP sythentase
Cold temps.
prevents tight packing/ soldification
Unsaturated tails needed
High temps.
prevents extreme fluidity
Down concentration
Across concentration
Small
Nonpolar
Down gradient
Contain
Coupled
Requires
Cell communication
Cell Singaling
Direct Contact
target
Local
long-distance
Pacarine
Synaptic
Hormone
Transcription
Reponse
Polar
Receptor
Cell Membrane
GPCR
Ion channel
TKR
Nnonpolar
Diffuse
Receptor
Cytoplasm
NUcleus
no singal
G protein inactive
Inactive enzyme
Signal
G protein active
GTP displaces GDP
active enzyme
Signal molecules
Reception
Dimerization
Phosphorylation
Ligand
Open
Kinase
Phosphate
Adds
Phosphatase
removes
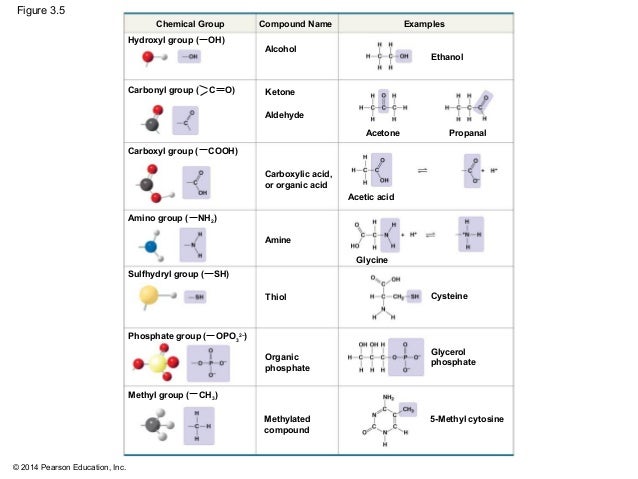
Functional Groups