Comparison of Prokaryotic and Eukaryotic DNA replication
Unlike Eukaryotes, which have advanced compartmentalization that allows them to have a nucleus, prokaryotes only have a nucleoid that stores their DNA.
Eukaryotic DNA is organized into several chromosomes consisting of a long, linear double-stranded DNA molecule that is helical. Prokaryotic DNA is one circle consisting of a double-stranded helical structure.
Both Eukaryotes and Prokaryotes have the same general process of initiation except prokaryotes only have one replication bubble.
Prokaryotic elongation has a similar process to eukaryotes for synthesizing lagging and leading strands. however, Prokaryotic elongation also includes the process of removing RNA primers and linking fragments.
Both Eukaryotic and Prokaryotic termination come from replication forks fusing. However, Eukaryotic replication forks fuse from 2 different replication bubbles, whereas Prokaryotic replicaiton forks that fuse come from the same replication bubble.
Process to make Protein
from mRNA
Amino acids
free ribosomes
The transcription process was already
in the cytoplasm
Terminator sequence is reached
and transcription stops
Complementary base pairing means that the nitrogenous bases can only bind with a specific molecule due to hydrogen bonding.
Adenine instead pairs with Uracil by forming 2 hydrogen bonds. Cytosine still binds with Guanine through the formation of 3 hydrogen bonds.
Adenine pairs with Thymine through the formation of 2 hydrogen bonds. Cytosine and Guanine form 3 hydrogen bonds.
Nucleotide
DNA Structure, Replication,
Expression, and Regulation
Structure of DNA and RNA
DNA
DNA is two connected polynucleotides (polymers) made up of nucleotides (monomer). The 2 sugar-phosphate backbones form a double helix.
Nitrogenous Bases
Purines are molecules consisting of 2 connected carbon rings. Purines include either Adenine (A) or Guanine (G) in both RNA and DNA.
Pyrimidines consist of a single carbon ring structure.
Thymine (T) and Cytosine (C)
Uracil (U) and Cytosine (C)
Sugar
Consists of pentose sugar called deoxyribose sugar which is missing a hydroxyl group on the 2' carbon.
Consists of pentose sugar called ribose sugar which includes the hydroxyl group on the 2' carbon.
Phosphate Group
Negatively charged phosphate groups link either deoxyribose sugar in DNA or ribose sugar in RNA through the formation of phosphodiester bonds.
Anti-parallel structure means that the two sugar-phosphate backbones run in opposite 5'-3' orientations.
DNA Replication
This model for DNA replication states the the resultant daughter molecules after replication consist each of one parental strand and one newly synthesized daughter strand.
DNA polymerase I and III
Initiation begins with a specific nucleotide sequence contained within the only ORI within the prokaryotic chromosome. A helicase binds to either template strand and begins expanding the replication bubble.
SSBPs will hydrogen bond to expoosed nitrogenous bases to prevent the dsDNA from reforming into a double helix.
Finally, Primase will lay down primers for DNA polymerase to expand into newly synthesized daughter strands of DNA.
As the replication bubble expands, the replication forks slowly move further away from the ORI. Synthesis of the lagging strand begins at an RNA primer near the replication fork on the 3' end of the template strand.
Elongation of the Okazaki fragment continues until the DNA polymerase encounters the RNA primer for the leading strand or the RNA primer for a previously synthesized okazaki fragment.
Elongation for the leading strand starts at RNA primers laid down at the ORI and is continuously synthesized from 5' to 3' in either direction towards the replication fork.
Elongation ends when DNA polymerase III runs into an already replicated portion of DNA.
The formation of 2 circular chromosomes, each of which consist of one parent strand and one newly synthesized daughter strand that are discontinuous and still connected.
At this point, the two daughter molecules are made continuous by nuclease first cleaving the phosphodiester bonds keeping the RNA primer in place. DNA polymerase I then adds DNA nucleotides where the Primer used to be, leaving a small gap until between it and the next strand.
DNA ligase removes the gaps by forming phosphodiester bonds between discontinuous daughter strands.
With the 2 new daughter molecules still attached, termination is the process by which the replication forks fuse thanks to recognizing certain termination sequences.
2 continuous circular chromosomes that are genetically equivalent and no longer connected.
2 helicase enzymes recognizes a specific nucleotide sequence called the origin of replication (ORI) and separates the double-stranded DNA (dsDNA) on either side of the replication bubble, forming 2 replication forks.
Topoisomerase helps relieve tension from the unwinding double helical structure of DNA as helicase widens the replication bubble.
As helicase moves in either direction away from the ORI, SSBPs hydrogen bond to the exposed nucleotides and prevent the dsDNA from reforming.
The enzyme primase will create an RNA primer which is elongated to allow DNA polymerase to bind and begin elongation.
The stage at which DNA polymerase extends the RNA primer or series of RNA primers with DNA nucleotides.
This strand starts at the ORI and is synthesized continuously towards the 5' end of the template strand or from the ORI towards the replication fork.
Starting at the RNA primer at the ORI, DNA polymerase III begins synthesis of the new daughter DNA strand continuously from the 3' end of the primer.
DNA and RNA can only be synthesized from 5' to 3', meaning the template strand is read from 3' to 5'.
Consists of a series of RNA primers going towards the 5' end of the template strand, but are synthesized from the replication fork towards the ORI in a series of Okazaki fragments. DNA polymerase then detaches before running into previous RNA primer.
As the replication bubble expands, Primase lays down an RNA primer near the replication fork at the 3' end of the template strand. As the primer is elongated by DNA polymerase III, the replication bubble continues to expand and a new RNA primer is laid down, which when elongated from 5' to the 3' will eventually end just before the RNA primer of the previous Okazaki fragment.
A series of short Okazaki fragments consisting of an RNA primer and an extended sequence of DNA nucleotides that aren't connected along the sugar-phosphate backbone.
Elongation ends when DNA polymerase III encounters sections of DNA that have already been replicated, leading it to disconnect from the DNA.
Another enzyme, nuclease, moves through the DNA and cleaves RNA primers. DNA polymerase then adds DNA nucleotides onto the daughter strand, leaving a small gap beetween where the RNA primer was and the next strand.
Termination ends with the final enzyme, DNA ligase, moving along the daughter strand and connecting the gaps between where RNA primers used to be by forming phosphodiester bonds.
Helicase
Untwists the double helix structure of DNA at the replication forks. Helps expose DNA to replicating machinery.
Topoisomerase
Enzyme responsible for relieving strain from unwinding the double helix.
Single-Strand Binding Proteins (SSBPs)
SSBPs help DNA replication by binding to the unwound parent DNA strands to prevent the hydrogen bonds from reforming.
Primase
Since DNA Polymerase can only add nucleotides onto an existing chain, primase creates a short RNA primer which then can be used by DNA polymerase to create a copy of the parent strand.
DNA Polymerase I and III
Adds nucleotides to the 3' end of RNA primer, detaching if/when it reaches the next primer.
Removes the RNA primer and replaces with DNA nucleotides.
DNA Ligase
Nuclease
Aids in DNA replication and repair by replacing certain damaged or incorrect sequences by cleaving the phosphodiester bond.
Helps join the final nucleotide between Okazaki fragments to make a continuous daughter strand through the formation of phosphodiester bonds in the sugar-phosphate backbone.
Protien Modification and Transport
Transcription and translation occurs in the cytoplasm, that is where the proteins are made
Prokaryotes only makes cells that they need , for that reason most are almost immediatly used
Translation occurs on free ribosomes, that is where proteins are created
Proteins leave the free ribosomes and goes to the ER
In the ER a carbohydrate group is added to the protein
The protein then travels from the ER to the Golgi
From the Golgi the proteins can move to other organelles/ locations with the help of secreting enzymes
Mitochondria, Chloroplast, Nucleus, Peroxisomes, cytoplasm
SRP (signal recognition particle)
First the SRP binds to the signal peptide in the free ribosomes, then SRP binds to a receptor protein located in the ER
Gene Regulation
Transcription and/or during mRNA processing
RNA Processing
Regulates by removing nucleotides that are not needed
The process of certain parts of the mRNA strand being removed to create the RNA strand that will be translated.
Enzyme: Spliceosome
Exons: nucleotides that stay in RNA to be used in translation to make a certain protein
because they exit the nucleus and go to the cytoplasm for translation
Intros: nucleotides that are not needed to make a certain protein
Stages of Transcription
Regulates by being specific with the nucleotides being added
occurs when the termination site is reached
A Poly-A Tail is added to the end of the mRNA strand
RNA nucleotides are added to the 3' end to create a chain of nucleotides
at some time during the process a 5' CAP is added to the mRNA strand
mRNA strand
Initiation
RNA Polyerase II binds to the promoter region to start transcription
A DNA strand is being used to make mRNA
the nucleus
Genes are regulated during transcription but Transcription and Translation occur simultaneously in the cytoplasm for prokaryotic cells.
Operons
Lac Operon
Lactose is present
The cell uses lactose to eventually make glucose and galactose for energy and carbon
The repressor will bind to lactose, preventing it from binding to the operator. This allows RNA polymerase to bind to the promoter to start transcription.
Lac A
Transacetylase
Adds acetyl group to lactose
Lac Y
Permease
Allows lactose to enter cell
Lac Z
Beta- Galactosidase
Breaks glycosidic linkages to form glucose and galactose
Glucose is Present
Lactose gene expression is off because the cell will rather use glucose for energy and for carbon.
Genes that can turn a function on or off
Operator
located next to the promoter
A protein will bind to the operator and either turn on or off gene trancription
Repressor
Turns off gene expression
Negative Regulation
Activator
Turns on gene expression
Positive Regulation
mRNA entering the cytoplasm
small ribosomal unit binding to 5' cap
Scans along the mRNA until
AUG sequnec is found,AKA starts codon
A tRNA with an anticodon of
3' UACS'
A tRNA molecule consist of a single RNA moluclue with about 80 nucletides
In three dimension tRNA
hasn anticodones on its ends
Large ribosome now
joins the complex
Has three active sites
P Site
Growing Polypeptide
E Site
Exist Site
A Site
Amino Acids
A translation initiation
complex is formed
Elongation
The next tRNA carrying the corrcet amino acid comes to site A
A peptide is formed between the two amino acids formed by the enzyme Peptidyl transferase
tRNA is now empty in the P site and moves to the E site to be released while the tRNA from A site moves to the P site and a new tRNA comes to the A site
mRNA is read from 5' to 3' direction while teh amino acids are added from N to C direction
Termination
Once a stop codon is reached in the A site no tRNA corresponds to a stop codon
A release factor sits in the A site
Dissociation of the ribosome complex causing translation to stop; This is a GTP driven process
Proteins called initiation factors
Hydrolysis of GTP provides
energy for the assembly
The initiator tRNA is in
the P site; the A site is available to the
tRNA bearing the next amino acid.
through hydrogen
bonds
Amino acid,Met
Transcription and RNA Processing
Synthesizing mRNA
In Cytoplasm for both Eukaryotic and
Prokaryotic Cells
Binding of transcription factors
Promotor Sequence
RNA Polymerase ll
Moves along the template DNA adding
ribonucleotides to the 3' end of the following RNA Strand
pre-mRNA
RNA Processing takes place
Splicesomes
Removes introns and splice together exons
mRNA
Prokaryotic cells
do have mRNA without the
slicing
3' end with a polyA tail
Enzyme called polyA polymerase
A 5' cap made of modified
guanine AKA cap or G-P-P-P
Stability
Translation
DNA sequences that
define where transcription of a gene by RNA polymerase begins
Activated adenylyl
cyclase converts ATP to cAMP; cAMP leads to cellular response
The C4 and CAM pathways are similar in that they both incorporate carbon dioxide into organic intermediates before entering the Calvin cycle. In C4 plants, carbon fixation and the Calvin cycle occur in different cells. In CAM plants, these processes occur in the same cells, but at different times of the day
In order to avoid photorespiration, some plants have adapted a different leaf anatomy where CO2 fixation and Calvin cycle occur in different cells. In C4 plants, CO2 is fixed in mesophyll cells and the Calvin cycle runs in separate bundle-sheath cells.
The most important of these are C4 photosynthesis and crassulacean acid metabolism (C A M)
Some plant species have evolved photosynthetic adaptations to minimize photorespiration in hot, arid climates
C3 Plants!
rubisco fixes O2 instead of C O2 and produces a two-carbon compound
Photorespiration consumes A T P, O2, and organic fuel from the Calvin cycle and releases C O2 without producing any ATP or sugar. Photorespiration interferes with CO2 fixation blocking photosynthesis. So this is not beneficial for the plant.
Photorespiration
6 CO2 + 18 ATP + 12 NADPH + 12 H2O
C6H12O6 + 18 ADP + 18 Pi + 12 NADP+ 6 O2 + 6 H2O + 12 H+
There are three phases to Calvin cycle – Phase 1 – Carbon fixation, Phase 2 – Reduction, Phase 3 – Regeneration of CO2 acceptor. The first step of phase I in Calvin cycle involves addition of CO2 from the atmosphere to Ribulose bisphosphate using an enzyme Rubisco (Ribulose bisphosphate carboxylase) (carbon fixation). This forms a 6 carbon unstable intermediate. This immediately splits to form two molecules of 3 carbon (3 phosphoglycerate). The figure shows 3 CO2 fixed one at a time, which means there are 6 molecules of 3 phosphoglycerate formed. Next step uses 6 molecules of ATP and then subsequently 6 molecules of NADPH. The product formed is 6 molecules of Glyceraldehyde 3 Phosphate (G3P), of which 5 molecules continue on to make more Ribulose bis phosphate (the carbon acceptor) and 1 molecule of G3P leaves the cycle to form glucose and other sugars.
ATP is made, NADPH (an electron carrier) is made and O2 is released. ATP and NADPH formed are used in the Calvin cycle to make sugars. Calvin cycle occurs in the stroma and used CO2 from the atmosphere.
In both cases energy generated from ETC is used to create a H+ gradient, which is then harvested to make ATP. So what is different between chemiosmosis in mitochondria and chloroplasts? Note first the differences in location where the H+ are pumped! There are also different components of ETC, but here we are more concerned about the difference in location.
Mitochondria and chloroplast are similar structures with similar evolutionary history. Therefore, both structures use the ETC to pump hydrogen out of the innermost membrane and use the resulting energy from H+ diffusion to drive ATP synthesis through ATP synthase.
Cellular respiration is generally the conversion of glucose and oxygen into water and carbon dioxide. Photosynthesis, however, does the opposite reaction, using water, light energy, and carbon dioxide in order to produce glucose.
In addition to chemiosmosis, cellular respiration makes ATP through substrate-level phosphorylation, whereas photosynthesis can make ATP through through cyclic electron flow in addition to chemiosmosis.
As electrons are transferred down the ETC, energy is released which is used to pump H+ against their concentration gradient (similar to respiration). H+s are pumped into the thylakoid space, so inside of thylakoid a lot H+ are present. These H+ get back out of the thylakoid down their concentration gradient. They do so by going through a protein called ATP synthase (just like in respiration). The energy associated with movement of H+ down concentration gradient is used to add an inorganic phosphate (Pi) group to ADP to form ATP. This mechanism of ATP formation is called Photophodphorylation
Light reactions and chemiosmosis: organization of thylakoid membrane
When there is excess NADPH present, the flow of electrons transiently shifts to a cyclic flow. Only PS1 is used, but no NADPH is formed. In this scenario, as electrons in PS1 are transferred to Fd, instead of forming NADPH they are recruited to the cytochrome complex and plastocyanin molecules of the electron transport chain. The movement of electrons leads to formation of ATP by photophosphorylation.
Photosystem I (PS I): The reaction-center chlorophyll a absorbs at 700 nm hence called P700
Phostosystem II (PS II): The reaction-center chlorophyll a absorbs at 680 nm hence called P680
A photosystem consists of a reaction-center complex surrounded by light-harvesting complexes
The light-harvesting complexes (pigment molecules bound to proteins) transfer the energy of photons to the reaction center. The reaction-center complex is an association of proteins holding a special pair of chlorophyll a molecules and a primary electron acceptor. In the reaction center, chlorophyll a transfers an excited electron to the primary electron acceptor. This redox reaction is one of the first steps of the light reactions
When pigments absorb light, an electron is elevated from a ground state to an unstable, excited state
Electrons fall back down to the ground state, releasing photons that cause an afterglow called fluorescence
If illuminated, an isolated solution of chlorophyll will fluoresce, giving off light and heat
Floating topic
Occurs in stroma
Occurs in thylakoid
A T P provides the necessary chemical energy, and N A D P H provides electrons needed to reduce C O2
C O2 is initially incorporated into an organic molecule through a process called carbon fixation
A T P is generated by adding a phosphate group to A D P in a process called photophosphorylation
The electron acceptor NADP+ is reduced to NADPH
O2 is released as a waste product
H2O is split to provide electrons and protons (H+)
Stage 2: The Calvin cycle produces sugar from C O2 with the help of the N A D P H and A T P produced by the light reactions
Stage 1: The light reactions convert solar energy into chemical energy
2 Stages
Electrons are extracted from water and transferred to CO2. So H20 is oxidized and CO2 is reduced
6 CO2 + 6H2O + Light energy = C6H12O6 + 6 O2
Stomata, which is where CO2 enters and O2 exits. These are microscopic openings on the surface of the leaf, and they can open or close accordingly.
Phosphodiesterase: An enzyme that catalyzes the hydrolysis of cAMP
Phosphorylation cascade
Uses Kinases and Phosphatases
Kinase remained inactive until
activated by addition of a phosphate group
Phosphate is removed from from a previous kinase which is describes as phosphatase
G Protein Coupled receptor (GPCR): Located on the membrane.
A signal molecule binds to the receptor, making the receptor change shape and activate
G Protein binds to GPCR which is then bound by GTP after replacing GDP; it does this by moving a phosphate group to the GDP attached to the G protein. This makes GDP into GTP and activates the G protein.
The G protein activates the enzyme; Adenylyl Case
can remove a phosphate
group= contains phosphatase function
Activated G protein activates enzyme and removes phosphate from GTP, converting to GDP
Adenylyl Cyclase: An enzyme located on the membrane that produces ATP by synthesizing ADP.
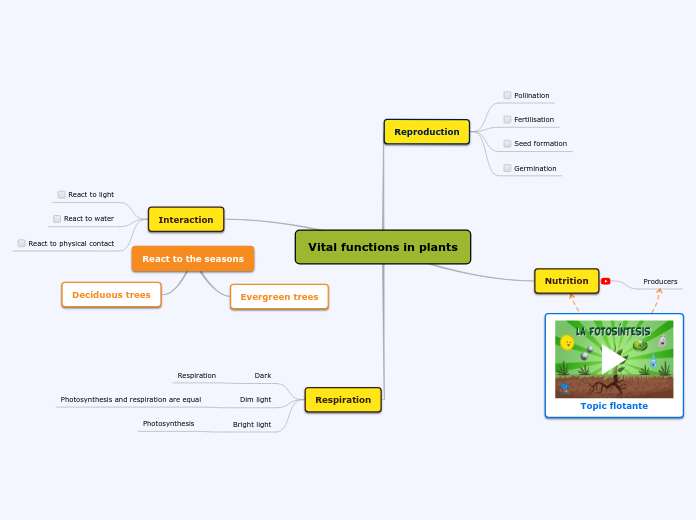
The main site of photosynthesis
About 30-40 chloroplasts
Specialized cells in the interior tissue of a leaf
Mesophyll
The process that plants use to create energy for food
Energy and Cell Communication
Cell Signaling
How cells communicate
By Releasing a signal
Target cell to have something to receive the signal molecule
Receptor
Intracellular Receptors
Intercellular Signaling
In cytoplasm, In nucleus
Nonpolar or hydrophobic
Steroid and Thyroid hormones
of animals and nitrite in
plants and animals
Membrane Receptors
Signal molecule is hydrophilic and can't
pass the membranes phospholipid bilayer; needs transmembrane recptores
Happens when signal molecule
is in the membrane;second messanger
Response
Signal triggers a certain
response
Transduction
Reception protein changes.
Multistep pathway to amplify
a signal
cAMP
Acts as second messanger in a G Protien
Signaling Pathway.
Activated adenylyl cyclase
converts ATP to cAMP
Binds to and activates another
protein, kinase, which goes to activate
another Kinase, leading to a cellular response
Reception
Chemical signal binds
to receptor protien
Types of membrane receptors
Tyrosine kinase receptor
Two pollypeptides that dimerize
Have the ability to function as a kinase
An enzyme that catalyze the transfer of phosphate groups from ATP to proteins
After phosphate group is added to the polypeptide after taken from ATP, the tyrosine is referred to as tyrosine kinase receptor
The activated receptor now interacts with other protiens to bring about cellular response
Ion Channel Recptor
Proton Pump
uses energy to transport protons from the matrix of the mitochondrion to the inter-membrane space
Its a proton generates a proton concentration gradient across the inner mitochondrial membrane because there are more protons outside the matrix than inside.
Sodium-Potassium Pump
Moves Na+ ions out of the cell and moves K+ ions into the cell. For every 3 Na+ ions pumped only 1 K+ is pumped.
Depolarization: Occurs when too may positive ions are pumped into the cell making the inside of the cell more positive than the outside.
Once Depolarization reaches a peak, repolarization occurs. It is the negative slope after the peak.
Repolarization: When the membrane potential is being changed back to a negative state.
Hyperpolarization: Occurs when too many positive ions are pumped out of the cell making the inside of the cell negative
Present in a target call that recovers the signal molecule
Molecules released by a cell to be recived by another cell
Long-distance
Hormonal Signaling
Local Signaling
Paracrine Signaling
Synaptic Signaling
Synapses are a chemical process that carries information from the presynptic
The arrival of action potential causes Ca2+
channels to open causing influx of Ca2+; the depolarization opens the voltage gates
Vesicles that were carrying neurotransmitters fuse with the presynaptic membrane and release them into the synaptic cleft
The neurotransmitter binds to
lingand-gated the ion channels in postsynaptic membrane.
Physical Contact
Extracellular Signaling
Occurs outside of the cell, usually signal molecules bind to a receptor located on the membrane.
G Protein linked receptor
Cell Respiration/ Aerobic Respiration
Anaerobic Respiration
Anaerobic Respiration is the process by which the body breaks down glucose without oxygen present. The body accomplishes this by using NADH as an electron acceptor instead of O2.
Glucose is converted into pyruvate through glycolysis, but instead of being oxidized, is reduced to regenerate NAD+ for use in the energy payoff phase of glycolysis to continually synthesize ATP through substrate-level phosphorylation.
Lactic Acid Fermentation
Alcohol Fermentation
Cellular respiration is the series of metabolic reactions that occur in cells in order to convert the chemical energy in food, in the presence of oxygen, to produce waste products in addition to adenosine triphosphate and other products to drive cellular work.
Glycolysis
2 Pyruvate, 2 NADH, and 2 ATP for each glucose.
Glycolysis is a sequential series of steps that occur in the cytosol of the cell that consist of an Energy Investment Phase and an Energy Payoff Phase in order to partially breakdown glucose and harness its chemical energy.
C6H12O6 + 2 NAD+ + 2 ADP + 2 P(i) --> 2 pyruvate (3-carbon) + 2 NADH + 2 H+ + 2 ATP
Pyruvate Oxidation
Refers to the process by which Pyruvate is transported to the mitochondrial matrix in eukaryotic cells, and oxidized into Acetyl CoA.
2 Pyruvate + 2 NAD+ --> 2 Acetyl CoA + 2 CO2 + 2 NADH + 2 H+
2 Pyruvate, 2 NAD+ + 2 H+
2 Acetyl CoA, 2 NADH, and 2 CO2
Citric Acid Cycle
The Citric Acid Cycle refers to the sequential series of 8 steps that fix Acetyl CoA onto oxaloacetate, where the repeated oxidation of this molecule produces intermediary molecules and the energy is captured by NAD+ or FAD+. This process is aided by enzymes at each step.
This phase, primarily characterized by the carbon fixation of Acetyl CoA onto oxaloacetate, involves steps 1 and 2. More specifically, this carbon fixation allows oxaloacetate to enter a higher energy state, then being oxidized.
This phase, including steps 3 and 4, involves the oxidation of isocitrate and the other intermediary molecules in order to reduce NAD+ into NADH, along with using substrate-level phosphorylation to produce ATP.
This phase, referencing step 5, includes the use of substrate-level phosphorylation in order to synthesize ATP.
This phase, including steps 6-8 involve the regeneration of oxaloacetate by oxidizing succinate and other intermediary molecules to reduce either NAD+ or FAD+.
Regenerated oxaloacetate, 2 NADH, and 2 FADH2.
2 ATP (one per Acetyl CoA)
you
2 Acetyl CoA
2 Isocitrate
2 Acetyl CoA + 6 NAD+ + 2 FAD+ + ADP + GTP --> 2 CO2 + 6 NADH + 6 H+ + 2 FADH2 + ATP + GDP
Oxidative Phosphorylation
Regenerates NAD+ and FAD+ in addition to producing 26-28 ATP molecules
Oxidative Phosphorylation is the final step in cellular respiration in which the chemical energy captured from food in NADH and FADH2 is moved through the electron transport chain to fuel the movement of protons out of the mitochondrial matrix and into the intermembrane space to drive the synthesis of ATP.
Electron Transport Chain/Respiratory Chain
Proton Gradient
Electrochemical Gradient
Chemical Gradient
The movement of molecules resulting from the uneven concentration of a solute in two different areas results in the passive diffusion of particles down the concentration gradient.
Electrical Gradient
The difference in membrane potential that results from the unequal concentration of ions on either side of a semi-permeable membrane. Drive ions away from areas of similar electric charge.
Energy-Coupling
Energy-coupling is the concept of coupling 2 biological reactions. This more specifically refers to the pairing of 2 biological reactions where one releases energy, and the other uses that energy to drive the second reaction.
The proton gradient is a biological "battery" that serves to store the potential chemical energy that was gathered from food and stored in NADH and FADH2.
Hydrogen exists in higher concentrations in the intermembrane space than in the matrix, meaning that region is also lower pH.
Protein Complexes 1, 3, and 4
Serve both as areas for redox reactions to take place for the Electron transport chain,
Protein Complex 1
NADH enters in the first protein complex because it has relatively higher potential energy than FADH2
Protein Complex 2
Cytochromes
Cytochromes are a group of proteins with heme bound that make up the majority of proteins in the ETC that donate and accept electrons.
FADH2 enters in the second protein complex because it has less potential energy than NADH, and thus enters at a lower energy level.
Chemiosmosis
ATP Synthase
ATP Synthase is the enzyme is chemiosmosis that is responsible for the facilitated diffusion of protons down their concentration gradient, which uses the energy from proton movement to synthesize ATP from ADP and inorganic phosphate.
Photosynthesis
Cytoplasmic
channels through cell walls
that connect the cytoplasms
of adjacent cells
Plasmodesmota
Organelles that have evolved to perform specialized functions in plant cells, including photosynthesis and the production and storage of metabolites.
Plastids
Cilia
Specialized arrangements of microtubules sheathed in a plasma membrane that are smaller and more numerous than flagella.
Can help with motility of the cell body.
The primary cilium, or a signal-receiving cilium, help transmit molecular signals from the environment to the interior of the cell.
Plasma Membrane
Composed primarily of phospholipids, membrane proteins such as cholesterol. Has a precise ratio of fats to phospholipids, and unsaturated to saturated, or degree of unsaturation, of lipids within the membrane.
Phospholipids consist of a hydrophobic head consisting of a phosphate group and a glycerol, connected to a 2 hydrocarbon tails.
The structure of the phospholipid means that in cells, the hydrophobic tails orient into a double layer with the hydrophobic heads facing the interior and exterior.
Higher saturation of the hydrocarbon tails in phospholipids will decrease membrane fluidity.
Membrane proteins, similar to phospholipids, orient with the nonpolar r-groups inside of the membrane.
Membrane proteins have a range of functions that include mediating the transport of ions and larger solutes through the membrane, helping with cell communication, and function as an enzyme to catalyze certain reactions.
Primary function of the Plasma Membrane is to separate the cytosol from the outside environment, selectively allowing passage of nutrients, water, and waste into and out of the cell.
Also plays an important role in anchoring the cytoskeleton to support cell shape.
More unsaturated carbons means greater means greater membrane fluidity.
Less Cholesterol present in the membrane results in greater membrane fluidity.
Higher temperature means greater membrane fluidity.
Takes up a significant amount of space in plant cells, serve as a repository for inorganic ions, including potassium and chloride, and maintains turgor pressure of the cell. All in all, it acts as a resevoir for water, a waste dump, a storage region, and maintainence of the cells shape.
Central Vacuole
Primary cell wall: relatively thin and flexible
Middle lamella: thin layer between primary walls of adjacent cells
Secondary cell wall (in some cells): added between the plasma membrane and the primary cell wall
Outer layer that maintains
cell’s shape and protects cell from
mechanical damage, it is responsible for plant cells rigidity.
Cellulose, as well as other polysaccharides, and protein
Cytoskeleton
Composed of actin filaments near the plasma membrane, intermediate filaments to anchor cell junctions, and microtubules to help organize the centrosome.
Organize microtubules to help with cell division, anchor cell junctions and organelles, support cell shape and structure, and help cells bind to the ECM.
Microtubules
Microfilaments
Intermediate Filaments
In between the size of microtubules and microfilaments, these structures are composed of several different types of proteins that are wound into tight coils.
helps maintain cell shape and anchor nucleus and other organelles.
Smallest of the 3, composed of 2 individual actin polymers wound together into a braid.
Helps with maintenance and changes in cell shape, muscle contractions, cell motility, and cell division.
Largest size of the 3 and composed of individual alpha and beta tubulin dimers that make up a polymer, wound into a hollow tube shape.
Helps with cell and organelle motility, maintenance of cell shape, and the movement of chromosomes in cell division.
Plant cells contain all of the same internal cell structures as those connected to animal cells, EXCEPT: centrosomes and centrioles.
Ribosomes and DNA, as well stacks of Thylakoid called granum are surrounded by aninternal fluid called stroma. These thylakoid are membranous sacs that contain chlorophyll.
Semi-permeable layer comprised
primarily of phospholipids. Separates the inner membrane from the cytosol of the cell.
b
2 distinct membranes which
are separated by an area known
as the inter membrane space.
Photosynthetic
organelle; converts energy of
sunlight to chemical energy
stored in sugar molecules
The Endosymbiont Theory describes
how the chloroplast was likely once a
photosynthetic prokaryote that was
engulfed into an earlier form of a eukaryote, eventually merging into a single
organism that we see today.
Chloroplasts
Inside the membrane
RNA Polymarse
a multi-unit enzyme that synthesizes RNA molecules from a template of DNA through a process called transcription.
One Kind
Several Kind
Archaeal translation is the process by which messenger RNA is translated into proteins in archaea.
Circular Chromosomes
Area where chromosomes are found
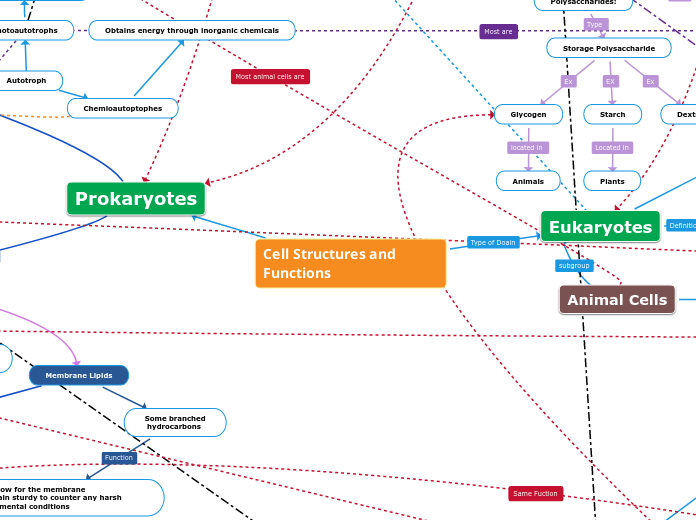
Heterotroph
Chemoheterotroph
Obtain energy from organic molecules
Photoheterotroph
Use light to obtain energy but CO2 is not the only carbon source
Autotroph
Chemioautoptophes
Obtains energy through inorganic chemicals
Photoautotrophs
Obtain energy from light to create their food
Endospores
Endospores are formed when
environmental conditions are unfavorable.
The original cell makes a copy of its chromosomes and surround them in a multilayer cell wall material
The cells are able to rehydrate
No membrane enclosed
organelles
Cell Structures and Functions
Eukaryotes
Any cell that contains a clearly defined nucleus and advanced compartmentalization of organelle activities.
The first single-celled organisms needed to have an organelle that supported both metabolic function and self-replication. Earliest organelle most likely to be ribozymes.
Thus, these early cells eventually consumed a oxygen using, energy-producing prokaryote to lead to the endosymbiont theory (see free ribosomes).
Ribozymes are a type of RNA with enzymatic function, helping jump start cellular life.
Plant Cells
Animal Cells
Internal Cell Structures
Nucleus
Nucleolus
Membrane-less structure primarily responsible for the synthesis of ribosome subunits and ribosomal RNA (rRNA)
Chromosome
Chromatin
Made up of proteins and nucleic acids.
Primary protein components are known as Histones.
Form into tightly wound coils to form chromosomes during cell division.
Discrete units of DNA within the nucleus that serve primarily to carry genetic information.
Nuclear Envelope
Double membrane each consisting of a phospholipid bilayer with embedded proteins.
Separates the nucleoplasm and cytoplasm, assisting in separating gene transcription from gene translation.
Runs continuous with Endoplasmic Reticulum and allow passage of molecules through protein structures in nuclear envelope known as nuclear pores.
Nuclear Lamina
Represents the inner face of the Nuclear Envelope and are involved in both gene expression (through chromatin organization) and helping to maintain structure of the nucleus.
Serves as the control center of the cell, housing most of the cell's DNA, made up of chromosomes, which guide gene expression and protein synthesis.
Nucleoplasm
Thick liquid that suspends contents of Nucleus.
External Cell Structures
Cellular Connections
Tight Junctions
Desmosomes
Gap Junctions
Consist of membrane proteins that sit in the membrane to allow passage of ions, sugars, amino acids, and other small molecules between cells.
Necessary for communication between cells in addition to the transport of molecules between cells.
Intermediate filaments made of keratin that anchor it into the cytoplasm and are responsible for connecting muscle cells in a muscle.
Helps fasten layers of cells together into a strong sheet.
Connect adjacent cells to allow the passage of some molecules (hence, "leaky" junction)
Consists of a network of proteins that interact to form connections from which no molecules can pass through.
Stops leakage of molecules within an environment into the outside environment or vice versa, allowing the body to compartmentalize more easily.
Tightly connects adjacent cells.
Flagella
Specialized arrangement of microtubules sheathed in the plasma membrane that project from some cells.
Primary function of the flagella is to help with the locomotion of the cell.
Extracellular Matrix
Composed of glycoproteins and other carb containing molecules.
The most abundant glycoprotein in animal cells, Collagen, is embedded in a network of proteoglycans. Here, fibronectin reacts with integrin in the ECM to bind cells to the surface.
Helps support the structural integrity and motility of the cell for functions such as amoeboid movement.
Provide Tracks for which transport proteins can transport molecules throughout the cell.
Cytoplasm
Thick, gel-like fluid that makes up all of the cell contained within the cell membrane.
Cytosol
Represents the intra-cellular fluid inside cells from which organelles are suspended.
Free Organelles
Free Ribosomes
Vacuole
Peroxisomes
Mitochondria
Centrosome
In certain animal cells, the centrosome resides near the nucleus and organize microtubules for the cytoskeleton and during cell division.
Centriole
A ring composed of microtubules arranged in nine sets of triplets.
A pair of centrioles oriented perpendicularly to each other compose a Centrosome
Consists of 2 phospholipid bilayers each with a unique set of membrane proteins.
The main site of cellular respiration in the cell, or the metabolic process that uses oxygen to metabolize macromolecules and convert that energy into ATP.
Outer membrane is smooth.
Separates the intermembrane space, or space between the inner and outer membrane, from the cytosol of the cell.
The inner membrane is convoluted with cristae, or folds in the inner membrane that help increase the surface area for sites of cellular respiration. Membrane proteins as enzymes help catalyze cellular respiration.
Separates intermembrane space from mitochondrial matrix, the compartment contained by the inner membrane.
The widely accepted endosymbiont theory states that early ancestors to eukaryotes engulfed oxygen-using, non-photosynthetic prokaryotic cells, forming a relationship between the prokaryote and the host.
This eventually led to the host cell and the endosymbiont merging into a single organism, a eukaryotic cell containing mitochondrion.
Specialized metabolic compartments bound by a single membrane with enzymes that remove hydrogen from certain molecules and transfer them to oxygen to make hydrogen peroxide (H2O2).
Detoxify harmful compounds such as alcohol by removing a hydrogen and making hydrogen peroxide.
Contain and dispose of toxic hydrogen peroxide.
Single membrane bound organelle that resembles a vesicle except for its larger size and longevity.
To help transport water, nutrients, and waste in and out of the cell.
Complexes made of ribosomal RNA and protein.
Sites where RNA and other cellular components come together to carry out protein synthesis.
Endomembrane System
Group of membranes and organelles that work together to metabolize, package, and transport lipids and proteins throughout the cell.
Lysosomes
Single membrane bound organelle containing a diverse set of enzymes.
Primarily responsible for the break down of macromolecules, worn-out cell parts, and pathogenic invaders for use by the cell.
Golgi Apparatus
Consists of Cisternae that separate each individual cisterna and the internal space from the cytosol.
Consists of Cis face ("receiving" side) and Trans face ("shipping" side)
Location in the cell where molecules are received, sorted, altered, or manufactured to be shipped out to other parts of the cell.
Vesicles
Compartment made of at least one lipid bilayer
Used to transport molecules in processes such as digestion and secretion.
Endoplasmic Reticulum
Consists of phospholipid bilayer with embedded proteins
Made of Cisternae, or interconnected, stacked, fluid-filled sacs, allowing the ER to run continuous through ER Lumen.
Rough Endoplasmic Retiuclum
Primarily involved in the steps of protein synthesis, including protein folding and protein sorting.
Contains Bounded Ribosomes
Ribosomes bound in the membrane of the Rough ER help with secretive protein and membrane-bound protein synthesis.
Smooth Endoplasmic Reticulum
Lacks Bounded Ribosomes.
Plays a key role in the synthesis of important lipids such as phospholipids and cholesterol.
Help breakdown drugs and other metabolic wastes
responsible for the production and secretion of some steroid hormones
Prokaryotes
Archaea
Peptidoglycan not present
in its Cell Wall
Extremophlies
When some archaea are
able to live in extreme
conditions
Extreme Halophiles
Live in highly saline
environments
Extreme Thermophiles
Thrive in very hot
environments
Membrane Lipids
Some branched
hydrocarbons
They allow for the membrane
to remain sturdy to counter any harsh environmental conditions
Bacteria
Nucleoid
Area where DNA is located because bacteria does NOT have a nucleus.
Exterior of Cell
Ribosomes
Capsule
Protects against dehydration
Polysaccharide layer
Carbohydrate
Sugars
Polysaccharides:
Structure Polysaccharide
Chitin
Cellulose
Plant Call Walls
Storage Polysaccharide
Dextran
Starch
Plants
Glycogen
Animals
Disaccharides
Glycosidic Bond/Linkage
Monosaccharides
Glyceraldehyde
Dihydroxyacetone
Ribulose
Ribose
Galactose
Fructose
Glucose
To provide energy
Cell Wall
It gives support, protects the cell, and maintains the cells shape.
Peptidoglycan
A polymer of modified sugars
cross-linked with short peptides
Gram Positive
Gram Negative
Fimbriae
Flagellum
Used to stick to
the substrate or to
another bacteria cell
Pili
Used for bacterial mating
Projections used to attach to surfaces