Cellular Processes
DNA Replication
Three Steps of DNA Replication
DNA polymerase I removes the RNA bases from the 5' end
The primer begins the leading strand. The existing strand is now the template strand. DNA polymerase III begins putting down the new bases. The lagging strand is synthesized in okazaki fragments. DNA ligase puts together the okazaki fragments.
Initiation
Replication begins at the ORI, where helices begins to unwind the DNA. Topoisomerase releases tension ahead of the replication fork. The RNA primers are created to begin elongation.
Structure of DNA
Complementary Bases
Cytosine
Guanine
Thymine
Adenine
Semi-Conservative Nature of DNA
DNA was discovered to be Semi-Conservative though the experiments by Messleson and Stahl. Both strands of parental molecule separate and each template synthesizes a new, complementary strand.
Cell Division
Cell Communication
Cells Signal one another frequently to tell other cells to begin replication, beginning at the phospholipid-bilayer.
This involves the Phospholipid-bilayer; where signal molecules activate cellular process, beginning with the attachment to a receptor.
DNA Synthesis
Origin of Replication
the point in the DNA at which replication begins
Replication Fork
point at which the two DNA strands begin to separate for replication
Lagging Strand
Synthesizes in a direction away from the fork
Leading Strand
Synthesizes in a direction toward the replication fork
Enzymes of DNA and their Functions
Topoisomerase
Prevents DNA from "overwinding" by releasing tension ahead of the replication
Ligase
Connects fragments of DNA
DNA Pol I
Removes RNA primer and replaces it with DNA
DNA Pol III
Synthesizes new DNA strands by adding nucleotides to 3' end of RNA primer
Primase
Creates RNA primer and places one at 5' end of leading strand and several on lagging strand
Helicase
Unwinds parental double helix at replication forks
Gene Expression
RNA Modifications after Transcription in Eukaryotic Cells
RNA Splicing
During RNA processing, the introns are cut out and the eons are spliced together.
5' Cap and Poly A Tail
A 5' cap and a Poly A tail are added to the two ends of a eukaryotic pre-mRNA molecule in order to 1.) facilitate the export of mature RNA from the nucleus 2.) protect the mRNA from degrading 3.)help ribosomes attach to the 5' end of the mRNA when it is in the cytoplasm.
3 Steps of Transcription
Termination
The RNA transcript is release and the polymerase detaches from the DNA
Elongation
The polymerase moves downstream, unwinding the DNA and elongating the RNA transcript in 5' to 3' direction.
1. The initiation of Transcription
RNA polymerase binds to the promote, the DNA strands unwind, and the polymerase initiates RNA synthesis at the start point on the template strand
Eukaryotes
3. Other transcription factors bind to the DNA along with RNA polymerase II, forming the transcription initiation complex.
2. Transcription factors bind to the DNA before RNA polymerase II can bind to it.
1. The eukaryotic promoter has a TATA box which is a nucleotide sequence containing TATA.
3. Termination of Tranlastion
3.The two ribosomal subunits and the other parts dissociate.
2. The release factor promotes hydrolysis of the bond between tRNA in P site and the last amino acid. Therefore, it frees the polypeptide from ribosome.
1. A ribosome reached the stop codon and the A site of the ribosome accepts a "release factor".
An rRNA molecule of a large ribosomal subunit catalyzes the formation of a peptide bond. This step removes the polypeptide from tRNA in the P site and attaches it to the amino acid on the tRNA in the A site.
1. The Initiation of Translation
2. A large ribosomal subunit arrives to complete the initiation complex. The initiation factors are in charge of connection all the translation components. Hydrolysis of GTP gives off energy to the assembly. The initiator tRNA is in the P site while the A site is free for the tRNA who carries the next amino acid.
1. A small ribosomal unit binds to an mRNA molecule. Then, an initiator tRNA, with anticodon UAC, pairs with the start codon, AUG, which carries the amino acid methionine (Met).
Translation
Molecule of mRNA is moved in a ribosome where codons are translates into amino acids. tRNA molecules interpret each type of nucleotide in sets of 3. A tRNA adds its amino acid to a growing polypeptide chain once the anticodon hydrogen bonds to the complementary codon on the mRNA.
2. Elongation Cycle of Translation
3. Translocation
Ribosome translocates tRNA in the A site to the P site. The empty tRNA in the P site is moved to the E site where it gets released. The mRNA moves with its bound tRNAs, which bring the next codon to the A site so it can be translated.
2. Peptide Bond Formation
1. Codon Recognition
Anticodon of aminoacyl tRNA base pairs with complementary mRNA codon in A site. Hydrolysis of GTP is present in this step. The aminoacyl tRNA with the appropriate anticodon binds and proceeds with the cycle.
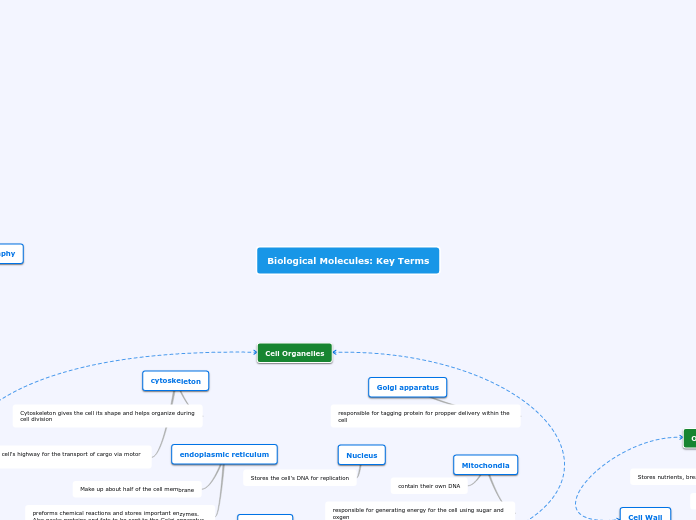
Cellular Components
Golgi Apparatus
Gives instructions to proteins and transports them
Endoplasmic Reticulum
Focuses on synthesizing protein (rough) and lipids (smooth).
Rough ER
Smooth ER
Ribosomes
Synthesizes proteins, made of RNA and proteins.
free or bound
proteins
Lysosome
A vesicle which breaks down
molecules and recycles them.
Endocytosis
Pinocytosis
Phagocytosis
Receptor Mediated
Nucleus
The component of a cell which
contains the genetic material of a cell.
Vacuole
Contains water or
various liquids in the cell.
Food Vacuole
Central Vacuole
Cytoplasm
An aqueous solution where the
cellular components reside in.
Cell Wall
A protective wall that is made
from cellulose and chitin
Mitochondria
Supplies energy to the cell through various cellular processes
Phospholipid Bilayer
A membrane made up of two fatty acid
tails and a hydrophillic head.
Glycerol
Fatty Acids (Fats)
Lipids
Present in plants, a layer of cellulose
and chitin which protects the cell.
Triglycerides (Fats)
Unsaturated
Oils
Saturated
Cell Signaling
Membrane Receptors
Ion channel receptor
A membrane-bound receptor which undergoes a conformational change once the signal molecule attaches to the receptor. Allows for the passage of ions.
Tyrosine kinase receptor
Allows for the transfer of phosphate from ATP to the tyrosine kinase regions of a dimer, once a signal molecule binds to it.
G protein coupled receptor
A cell surface transmembrane receptor that works with the help of a G protein.
Types of Signal Molecules
Receptor on/in Cell (Plasma) Membrane
Hydrophobic Signal Molecule
Receptor in Cell Nucleus
Receptor in Cell Cytoplasm
Types of Cell Signaling
Long-Distance Signaling
Exocrine/Hormonal Signaling
Hormone must travel through the blood stream
Local Signaling
Synaptic Signaling
Neurotransmitters released which diffuses across the synapse, stimulating the target cell.
Paracrine Signaling
Involves direct signaling: affects nearby cells
3 Stages of Cell Signaling
Cellular Response: The transduced signal triggers a cellular response
Transduction: The receptor protein changes in some kind of way when the signaling molecule binds to it. The signal is conveyed to a form that can bring about a specific cellular response. Transduction usually occurs in a series of steps-signal transduction pathway.
AKA: Amplification (involves Second Messengers)
Reception: A signal molecule binds to a receptor protein located at the cell's surface.
Gene Regulation
Transcription Factors
Specific transcription ,like activators, lead to increased expression (high levels of transcription)
Distal control elements are enhancers that are located upstream or downstream of a gene
Activators bind to the enhancer sequence and bends the sequence
General transcription factors leads to low(basal) levels of transcription
Proximal control elements are sequences in DNA close to promoter
Transcription factors binds to the proximal elements and transcription is low level
allows RNA pol II to bind weakly
Repressor binds to the enhancer sequence and bends but prevents RNA poly II to bind
Tryptophan
When tryptophan is present, the repressor is active which means it can bind to the operator, operon is off
When tryptophan is absent, the repressor is inactivated which means it can't bind to the operator,operon is on
A negative regulator because it binds to a repressor
an amino acid
cAMP Levels
Lactose is present, glucose is present, cAMP levels are low and CAP is inactivated
Lactose is present, glucose is absent, cAMP levels are high and the CAP is activated
Operon is on!
Lactose is present, repressor is inactive(allolactose inactive the repressor), operon on
Operon is off!
Lactose is absent, repressor is on(repressor binds to the operator), opreron is off
Lac Operon
Lactose operon is an operon required for the transport and metabolism of lactose in bacteria.
Lac I is the lac repressor (regulatory gene)
Can switch off the operon when binding to the operator and it is active by itself.
Lac A adds an acidicyl group to lactose
Enzyme responsible is transacetylase
Lac Y takes in the lactose
Enzymes responsible is permase
Lac Z is a gene that forms mRNA
Enzymes responsible is galactosidase
Prokaryotes
Operator is not in Eukaryotes it is in Prokarytoes
Multiple genes are controlled by the same promoter.
In prokaryotes, genes are organized as operons.
DNA Packaging
The DNA is packaged with proteins in an elaborate complex known as chromatin (the basic unit of the nucleosome).
Histone core: a protein that bind the DNA twice
Nuclesomes are formed when the DNA wraps around the histones twice.
The looped domains coil further.
The 30-nm fiber forms looped domains that attach to proteins.
Interactions between nucleosomes cause the thin fiber to coil or fold into this thicker fiber.
The tight helical fiber needs help of the H1 which is outside the nucleosome.
Nuclesomes are strung together like beads on a string by linker DNA.
Only the H2A, H2B, H3, H4 form the histone core. The H1 is outside and not apart of the code.
Different types of Histones: H1, H2A, H2B, H3, H4
Think of "beads on a string"