central dogma of molecular biology
a theory stating that genetic information only flows one direction
RNA directly to protein
DNA to RNA to Protein
DNA is transferred to mRNA molecule by a process called transcription
mRNA is bound by ribosomes which read mRNA to produce chain of amino acids
DNA Structure, Replication, Expression and Regulation
endomembrane system
membranes and organelles in eukaryotic cells
package, transport, and export the proteins made in cell
composed of nuclear envelope, endoplasmic reticulum, Golgi Apparatus, vesicles, lysosomes, and vacuoles.
protein could either function as membrane protein or be secreted outside of the cell
regulation
both prokaryotic & eukaryotic cells have the ability to regulate their gene expression
prokaryotes need to often change their metabolic pathways due the availability of nutrients, which can be done by regulating expression of certain genes
most common way they regulate genes is through the use of operons
operon: a group of prokaryotic genes of related function which are controlled by a single promoter
lac operon: inducible operon with 3 genes encoding enzymes that metabolize lactose for energy; only in the presence of lactose (& absence of glucose) is the lac operon transcribed
glucose impact on lac operon:
glucose is the preferred energy source even in the presence of lactose in most prokaryotes
if glucose is available, the lac operon should be turned "off"
glucose levels are linked to cyclic AMP (cAMP)
when glucose is low/absent. cellular levels of cAMP increases
high cellular cAMP levels increases the rate of transcription of the lac operon
Lac A
Lac Y
Lac Z
operator: regulates transcription of the operon; decides whether the gene should be "off" or "on" through the binding of regulatory proteins
regulatory proteins: binds to the operator and affects RNA polymerase binding to the promoter
repressor: blocks RNA polymerase binding
(prevents transcription)
Lac I: active repressor protein that normally represses transcription when bound to lac operator
activator: promotes RNA polymerase binding
(stimulates transcription)
promoter: a region of DNA that initates transcripiton of a gene
it is located at the beginning of the gene
gene expression: the ability to express a gene to produce products such as proteins and non-coding RNA molecules
regulated in 2 ways:
negative regulation
prevents gene expression by turning "off" the gene
positive regulation
stimulates gene expression by turning "on" the gene; genes are allowed to be expressed
differential gene expression: process that allows multi-cellular orgnaism to express genes differently in different cells
all cells of a multi-cellular organism have the same genome, but have different sets of proteins
fundamental to eukaryotes
can be controlled at any of these 5 stages:
post-translational control: regulates modifications to proteins after translation
can activate/inactivate a protein for degradation by proteases
proteases: enzyme that degrades proteins by breaking polypeptide bonds making single amino acids
translational control: regulates initiation and elongation steps of translation
post-transcriptional control: regulates modifications to RNA after transcription
small noncoding RNA block translation of target mRNA molecules
ribosome is blocked from binding
mRNA is degraded
RNA processing adds 5' Cap & poly-A tail to mRNA for protection from RNA degrading enzymes in the cytoplasm
alternative RNA splicing
results in different protein products form the same mRNA transcript
spliceosome: the RNA protein complex that removes introns from premature RNA
introns: noncoding section of an RNA trancription and interrupts the sequence of genes
transcriptional control: regulates RNA polymerase binding to a promoter & initiation of transcription
transcription intiation in Eukaryotes requires a complex of trancription factors bound to the promoter sequence so then RNA pol II can bind
transcription factors: proteins that bind to specific DNA sequences & regulates transcription initiation
specific: bind to distal control elements called enhancers
repressors: brings about low levels of transcription
activators: bring about increased level of transcription
general: bind to (or near) the promoter
recruits RNA polymerase to the promoter region of a gene
brings about low levels of transcription (background/basal)
recruited by the TATA box
TATA box sequence of A & T repeats located in the promoter
most prokaryotic gene regulation occurs at this stage
chromatin rearrangements: regulates chromatin conformation & DNA's accessibility for transcription
chromatin: loosely coiled nucleosomes
nucleosomes: DNA wrapped around units of 8 histone proteins (octamer)
histones: proteins that binds the DNA (first level of packaging
H4
H3
H2B
H2A
H1
it is NOT involed in the octamer (it acts independently)
DNA STRUCTURE
a double helix formed from two complementary strands of nucleotides held together by hydrogen bonds between G-C and A-T base pairs.
DNA strand is used as template strand to create new complementary strand
cytosine, guanine, thymine, adenine
Replication mechanism
monomer
nucleotides
hydrogen bonds connect complementary nucleotides
three major steps
assemply of new DNA segment
priming of template strands
opening of the double helix and separation of the strands
Membranes,
Energy,
and Cell Communication
Cell membranes
types of membrane transport
active transport
simple passive
selectively permeable
Phospholipids
basic component
phosphate
2 fatty acids
glycerol
cell communication
transduction
second messenger
cyclic AMP (cAMP)
converted to AMP by phosphodiesterase
formed from ATP using Adenylyl Cyclase
two types of receptors
intracellular receptors
membrane receptors
Ion Channel receptor
Tyrosine Kinase receptor
made of two polypeptides which dimerize when a signal molecule is bound to each polypeptide
each polypeptide takes a phosphate group from ATP and adds it to the other polypeptide
called autophosphorylation
G protein linked receptor
steps at reception:
signal molecule binds to GPCR
allows G protein to bind
causes GDP to be replaced with GTP
active G protein activates enzyme
*G protein switch removes phosphate group from GTP to make GDP
sending and receives signals
long distance signaling
hormonal signaling
local signaling
paracrine & synaptic
Energy
it is constantly being changed from one type of energy to another
thermodynamics: the study of energy transformations
second law: every energy transfer or transformation increases the entropy of the universe
entropy (S): measure of disorder
Gibbs free energy (G): helps to predict the spontaneity (or lack thereof) of a reaction at constant temperature and pressure
ΔG=ΔH-TΔS
ΔG>0 implies that ΔStotal<0
ΔG=0 implies that ΔStotal=0
ΔG<0 implies that ΔStotal>0
first law of thermodynamics: energy can be transferred or transformed but it cannot be created nor destroyed
surroundings: matter in the rest of the universe
system: the matter under study
open system
closed system
e.g. photosynthesis: when light energy (kinetic energy)is converted into chemical energy (potential energy) to transform it into glucose
broken up into two stages
calvin cycle
broken up into three stages
RuBP Regeneration
a series of enzymatic reactions driven by ATP
G3P synthesis
gl
carbon fixation
catalyzed by enzyme Rubisco
uses CO2 & chemical energy to synthesize glucose
light reactions
occurs in the thylakoid membrane/space
converts light (photons) & H2O into chemical energy (ATP & NADPH) while producing O2 as a byproduct.
chemical energy will be used to power the Calvin Cycle
ATP: energy currency of the cell
different ways to synthesize ATP in cells
Oxidative phosphorylation: electrons from NADH and FADH2 transfer to oxygen, generating ATP; occurs in the mitochondria
chemiosomosis
following ETC, so many protons are now being pumped into the intermembrane space, and they now want to go back their concentration gradient into the matrix trhough facilitated diffusion.
facilitated diffusion occurs through the used of the enzyme, ATP synthase. This results in the synthesis of ATP
26-28 ATP
electron transport chain (ETC)
NADH brings electrons from glycolysis and citric acid cycle where they move down complexes 1, Q, 2,3, cyctochrome c, 4, and finally oxygen to form water. While this is occuring, there is a release of energy.
the energy produced is used to pump H+ against their concentration gradient (proton gradient) into the intermembrane space
an example of active transport
substrate level phosphorylation: when a phosphorylated substrate and ADP interacts with an enzyme which leads to a formation of a product (substrate) and ATP (when the phosphate group from the substarte is transferred to ATP)
citric acid cycle
following glycolysis, the two pyruvate formed are oxidized to form acetyl-Coa, our starting molecule.
molecules per acetyl-Coa formed: (there are 2 acetyl-Coa present! so each molecule times two)
ATP: 1
NADH: 3
FADH2:1
glycolysis
you start with glucose and ATP (2)
energy payoff phase: the last five steps you make more ATP than you used
net number of molecules:
ATP: 2 (2 used and 4 formed)
NADH: 2
Pyruvate: 2
energy investment: the first five steps you use ATP
occurs in the cytoplasm (outside the mitochondria so it does NOT need oxygen; also known as anaerobic respiration)
renewable resource regenerated by the addition of a phosphate group to ADP
catabolic reactions in the cell power the phosphorylation of ADP
NADPH: electron donor
6CO2 +6H2O -> C6H12O6 +6O2
products are provided to the mitochondria
goes through cellular respiration
C6H12O6+6O2 -> 6CO2+6H2O+ATP
anaerobic respiration: occurs without oxygen and releases energy quickly
lactic acid fermentation: pyruvate (2) is directly reduced, grabbing electrons from NADH, forming lactate (2)
humans can do this
alcoholic fermentation: a pyruvate (2) forms acetyl-dehyde which gets reduced with the electrons from NADH forming alcohol
humans can't do this
aerobic respiration: occurs with oxygen and releases energy slowly
C6H12O6+6O2 -> 6CO2+6H2O+ATP
products (minus ATP!) are used as reactants for photosynthesis
the ability to do work
it is used, stored, and transformed in living systems
metabolism: the totality of the chemical reactions of an organism's body
chemical reactions can be broken down into two pathways
anabolic
nonspontaneous
endergonic
when simpler molecules are converted into complex molecules, consuming energy
e.g. photosynthesis
catabolic
spontaneous
exergonic
breaking down complex molecules into simpler compounds, releasing energy
e.g. cell respiration
a starting molecule is converted into a product through the used of intermediates
catalyzed by specific enzymes suited for the reaction
enzymes help to lower the energy barrier which reactant need to overcome before they can form products
regulation of enzyme function
allosteric regulation
regulatory molecule binds to a protein
at one site which then affects the protein's function at another site
regulatory molecule
activator: stimulates enzyme activity
e.g. cooperativity: when one substrate molecule binds to an active sits allowing all other subunits to go into active form
inhibitor: inhibits enzyme activity
inhibition of enzyme activity
noncompetitive inhibition: a noncompetitive inhibitor binds to the enzyme away from the active site, changing its shape so that the active site functions much less effectively
feedback inhibition: when the end product of a process stops the process from continuing
competitive inhibition: a competitive inhibitor mimics a substrate, competing for the active site
substrate: the small molecule that an enzyme binds to
binding of a susbtrate forms weak bonds, changing the shape of the enzyme
weak bonds include hydrogen bonds and ionic bonds
active site: where on the enzyme the substrate binds to
Potential Energy
stored energy available to do work
e.g gravitational energy
e.g. chemical energy
Kinetic Energy
energy of motion
e.g. muscle contractions
e.g. light energy
membrane proteins
basic
acidic
nonpolar
types of R groups present
polar
chemical bonds, cell structure and functions
Linkage
shared electrons=pair of electrons
the chemical bond is composed by 2 electrons coming from the outer layer of each different atom to make a pair of electronds
chemical bond is a link between 2 atoms to give a molecule
provides energy necessary to form a chemical
strength of the bond depends on the molecules involved in the process of bond formation
biological importance
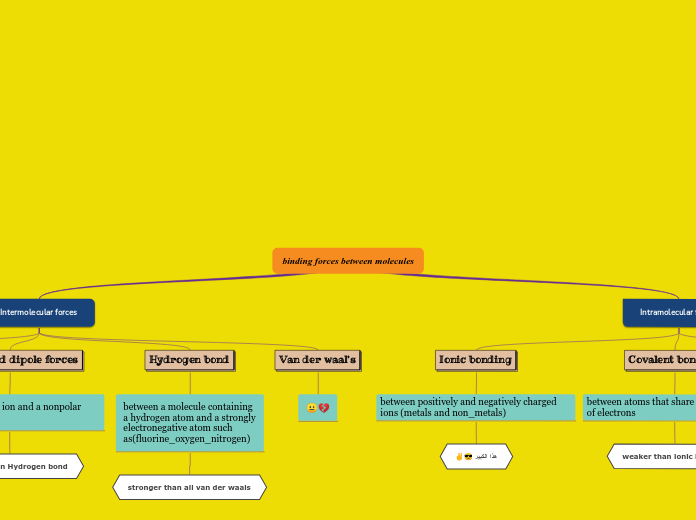
hydrogen
makes water molecules stick together. responsible of the properties of water.
cause protein chains to spiral and bend, giving unique shapes
ionic
compounds with ionic bonds split into ions in water. Ions conduct electricity. Gives specialized cells excitable properties
covalent
holds together the long chains of macromolecules (DNA, RNA, and Proteins)
Chemical bonds
Glycosidic bond
type of covalent bond that joins a sugar molecule to another group which could be another carbohydrate.
Phosphodiester bond
make up backbone strands of DNA and RNA
this bond is the linkage between the 3" carbon atom of one sugar molecule and the 5" carbon atom of another
these bonds are central to all life on earth
Peptide bond
proteins are linear polymers composed of amino acids linked by a peptide bond
chains containing less than 50 amino acids are peptides
chains containing greater than 50 amino acids are called proteins
peptide bonds are formed by the condensation of the carboxyl group of amino acid and the amino group of the second amino acid with the elimination of water
Metallic bonds
type of bonding found in metallic elements
electrostatic force of attraction between positively charged ions and delocalized outer electrons
refers to an interaction between delocalized electrons and the metal nuclei
Example of metallic bonding: if metal cations and electrons are oppositely charged they will be attracted to each other and also other metal cations
Hydrogen bonds
attractive force between the hydrogen attached to an electronegative atom of one molecule and one from a different molecule
the electronegative atom is usually oxygen, nitrogen, or fluorine.
these have partial negative charges
example of hydrogen bond: a hydrogen atom covalently bonded to an oxygen via a shared pair of electrons
Ion- dipole
attractive forces between polar molecules and ions
Ionic bonds
attraction between ions of opposite charges
complete transfer of valence electrons between atoms
metal loses electrons to become a positively charged cation
non metal accepts these electrons to become a negatively charged anion
example of ionic bond: Na and Cl
Covalent bonds
two types:
Add your text
nonpolar covalent
evenly matched
polar covalent
unevenly matched, but willing to share
example of covalent bond: two hydrogen bonds getting close together, the attraction is balanced in both directions. Hydrogen gas is formed.
strongest bond: sharing of electron pairs
biological macromolecules
Lipids
not a true polymer
waxes
fatty acids bound to long chain alcohol molecules
functions:
prevention of water loss
protection
steroids
essential for the structure of
animal cell membranes
made up of 4 fused carbon ring structures
triglycerides
a lipid with 3 fatty acid chains
linked to a glycerol molecule
occurs through a dehydration reaction
phospholipids
has two fatty acid chains
otherwise known as the "tail"
hydrophobic
composes all cell membranes
contains a phosphate group
otherwise known as the "head"
hydrophilic
fatty acids
unsaturated
not fully saturated with hydrogens
double bonds
causes kinks in the chain
liquid at room temperature
saturated
saturated with hydrogen
single bonds
able to pack closely together
higher melting points
solid at room temperatue
Carbohydrates
carbon-based molecules hydrated with many hydroxyl groups (-OH)
monomers: monosaccharides
polymers: polysaccharides
types:
complex carbohydrates
starch
composed of alpha glucose molecules
can be digest by humans due to the
amylase enzyme
cellulose
composed of beta glucose molecules
cannot be digest by humans because we
lack the necessary enzyme needed to break it down
simple carbohydrates
most abudant is glucose
C6H12O6
bonded through glycosidic linkages
form via a dehydration reaction
Nucleic Acids
monomer: nucleotide
phosphate group
sugar molecule
nitrogenous bases
purines
guanine
adenine
pyrimidines
uracil
thymine
cytosine
polymers:
DNA
forms a double helix
with anti-parallel strands
connected by base-pair hydrogen bonding
sugar molecule: deoxyribose
thymine (T)
RNA
forms a single-stranded nucleotide chain
sugar molecule: ribose
base pairs:
guanine (G)
cytosine (C)
uracil (U)
adenine (A)
has directionality (5' & 3' ends)
bonded through phosphodiester bonds
results in the sugar-phopshate backbone
is formed through dehydration synthesis
also known as condensation reaction
Proteins
monomers: amino acids
20 different amino acids are used by living organisms
protein polymers
Structure and Organization
Quartenary: arrangement of multiple polypetide chains to form a protein
Tertiary: 3D-shape of polypeptide chain
determined by R group interactions
Secondary: formation of α helices or β pleated sheets
occurs due to the formation of hydrogen bonds
Primary: types, quantity, and sequence of amino acids
has directionality (N- terminal & C-terminal ends)
bonded through peptide bonds
cells
chemical evolution hypothesis
three domains of life
archaea
methanogens live in swamps and produce methane as a waste product
strict anaerobes
extreme thermophiles: very hot environments
extreme halophiles: live in saline environments
branched membrane lipids
eukaria
made up of eukaryotes
most of the DNA is in the nucleus
nucleus is an organelle that is bounded by a double membrane
cells have membrane bound nucleus
bacteria
c
DNA in nucleoid
no membrane bound organelles
membranes
membrane proteins
functions
attachment to ECM an cytoskeleton
intercellular joining
cell-cell recognition
signal transduction
enzymatic actvity
transport
active transport
specific case of active transport is the sodium-potassium pump
uses energy
maintains a concentration gradient
movement of substances from low to high concentrations
passive transoport
facilitated diffusion
passive transport aided by proteins
example includes osmosis
water balance of cells
hypotonic
solute concentration is less than that inside the cell; cell gains water
hypertonic
solute concentration is greater than that inside the cell; cell loses water
isotonic
solute concentration is the same as inside the cell; no net water movement across the plasma membrane
tonicity
ability of a surrounding solution to cause a cell to gain or lose water
diffusion of a substance across a membrane with no energy investment
membrane fluidity
Each phospholipid has a specific temp
temp affects the fluidity
below temp lipid is in gel phase and is rigid
above temp lipid is in liquid crystalline phase and is fluid
plasma membrane
regulate cell's tarffic
consists of phospholipid bilayer that is semipermeable
hydrophobic fatty acid tail (away from water) and a hydrophilic head (faces water)
hydrophilic becuase of phophate group
amphipathic
cholesterol
no nucleus
membrane transport
Phospholipids and proteins
make up most of the membrane
cholesterol helps with flexibility and carbohydrate chains help communicate with other cells
phospholipid: 2 fatty acid tails and a phosphate head
phospholipid bilayer forms because the inside and outside of the cell are mostly water
semipermeable because it only allows certain molecules to cross
Hydrophilic, polar, large and charged molecules must use a transport Protein to enter/exit the cell.
transport proteins
There are two types of Transport Proteins: �Protein Channels: a special entryway for large, polar, hydrophilic and charged ions to diffuse through the cell membrane�This is called Facilitated Diffusion.
The proteins are specific; glucose can only pass through a glucose transport protein.
The fluid mosaic model
the membrane is made up of many smaller parts and the structure moves likes a fluid
Semipermeable membranes
membranes that allow certain materials to pass through based on certain properties
size, hydrophobicity, charge
The cell boundary
seperates cellular materials from external environment
regulates which materials can enter and exit the cell
maintains homeostasis in cell
membranes are found around the cell and each organelle