jonka Joseph-Hoang Nguyen 8 kuukautta sitten
274
h
jonka Joseph-Hoang Nguyen 8 kuukautta sitten
274

Lisää tämän kaltaisia
G-proteins act as molecular switches in the signaling pathways initiated by receptors like GPCRs (G-protein-coupled receptors).
Uses electrons from PSII
Makes NADPH
When there is too much NADPH, changes to cyclic flow.
"Catch-up" on making ATP.
Products of Non-cyclic:
H2O Used
O2 Released through Stomata
Electrons used from H2O
ATP Made
Outputs: Sugar, NADP, ADP
Inputs: CO2, ATP, NADPH
Produces sugar from CO2
Location: Stoma
Phase 1: Carbon Fixation
CO2
(+ rubisco): 6C (Short term intermediate, unstable)
3-Phosphoglycerate
(+ 6 Phosphates): 1,3-Bisphoglycerate
(- 6 phosphate due to NADPH): Glyceraldehyde-3-phosphate (G3P)
One G3P leaves, makes sugar, rest back to regen RuBp
Outputs: ATP, NADPH, O2
Inputs: Light, ADP, NADP+, H2O
Solar Energy --> Chemical Energy
Location: Thylakoid Space
Photosystems
Hetero and Autotrophs (Individual)
Metabolic rate how much energy an organism uses, affecting energy needs.
Energy Flow in Populations: Individual energy needs impact group energy consumption.
Energy Loss Between Levels (Community): Only about 10% of energy is passed up each trophic level; the rest is lost as heat.
Cells break down glucose with oxygen to release energy, CO₂, and water.
ATP Production: Energy from respiration is stored in ATP molecules for cellular functions
Energy Loss as Heat: Not all energy is stored; some is lost as heat during metabolic processes.
Requires Oxygen
Glycolosis
1st Step:
Glucose to Glucose-6-Phosphate with the enzyme hexokinase.
3rd Step:
Fructose-6-Phosphate to fructose-1-6-phosphate with the enzyme phosphofructokinase.
Output:
2 Net ATP
2 Pyruvate
2 NADH
Pyruvate Oxidation
Pyruvate Oxidation requires oxygen.
Pyruvate makes:
1 Acetyl CoA
1 NADH
Citric Acid Cycle
Citric Acid Cycle (Krebs Cycle/TCA Cycle)
Step 1:
Acetyl CoA and Oxaloacetate go together and make Citrate.
Step 3:
Isocitrate becomes ketoglutarate
Output:
1 ATP
3 NADH
1 FADH2
Oxidative Phospohlation
This process is broken down into two parts, the Electron Transport Chain(ETC) and Chemiosmosis.
Chemiosmosis
Once many H+ exits into the intramitochondrial space through the ETC, ATP synthase allows H+ to go back into the matrix in through facilitated diffusion in attempt to even out on both sides. This takes ADP turning it into ATP.
Electron Transport Chain
This process includes:
Complex I
Complex II
Complex III
Complex IV
Complex Q
Cyc
NADH gives an electron to complex one turning it into NAD+. FADH2 gives electron to Complex 2, turning into FAD. They both give their electrons to Q, from Q to complex III, Complex III to Cyc, then to complex IV and given to oxygen where it makes water(H2O). Complexes I, III, and IV are proton pump which pump out H+ into the intramitochondrial space when the electrons are being passed around.
Doesn't Require Oxygen
Alcohol Fermentation
When there is no O2, pyruvate forms acetaldehyde and is reduced to ethanol where CO2 is released.
This reduced electrons from NADH allowing glycolysis to continue.
Lactic Fermentation
When oxygen is not present, pyruvate is reduced and forms lactate and recycled back into NAD+ allowing glycolysis to continue. In Lactic Acid fermentation CO2 is not formed.
Each phospholipid has a specific phase transition temperature.
Above this temperature, the lipid is a fluid and it is in its liquid crystalline phase.
Below this temperature, the lipid is rigid and in a gel phase.
A thin, semi-permeable membrane that separates the inside of a cell from the outside environment.
Hydrophobic tail
Creates selectively permeable membrane
Controls what substances can pass through the cell
Repels water
Hydrophilic head
Attracts water into the membrane
Contransport
Proton Pump
Electrogenic Pumps
Ion Channels
Sodium-Potassium Pump
Osmosis
Diffusion
Facilitated Diffusion
Local
Synaptic Signalling
Paracrine signalling
Long Distance
Hormal Signalling
Plants
Animals
Tight Junctions
Gap Junctions
Desmosomes
Breaking down of molecules and defective components
The take in of nutrients to produce ATP through cellular respiration
Mitochondria
m
Active Transport
Carried out by the Cell/Plasma Membrane
Passive Transport
Cytoskeleton (Plant & Animal Cells)
Cell Wall (Plant Cells)
A chemical bond refers to a force of attraction between two or more atoms that are held together to form molecules.
Atoms share electrons in covalent bonds.
Intramolecular
Bonds between atoms in molecules.
Polar Covalent Bonds
Hydrogen Bonds
Van der Waals Forces
Interactions of electrons of nonpolar susbtances.
Hydrogen will have a slight positive charge, so it will be attracted to neighboring negative charges.
Electrons are unequally shared by atoms.
Electrons spend more time close to one atom rather than another.
Partial Negative Charge
Partial Positive Charge
Nonpolar Covalent Bonds
Two atoms share electrons somewhat equally.
Ions are charged particles.
Cations
Positive ions formed by losing electrons.
Anions
Negative ions are formed by electron gain.
Ionic Bonds
Bonds formed between ions with opposite charges.
Pilli/Fimbre
Pilli are hairs that help in movement
Fimbre stick out and can attach to stuff
Capsules
Nucleoid
Area where chromosomes are
Cytoplasm
Filling in the cell/hold organelles/components in place
Ribosomes
Synthesize Proteins
Plasma Membrane
Separated the outside and inside of a cell and controls what goes in and out.
Capsule/Slime Layer
Layer of Protection
Flagella
Aids in movement
Vesicles
Used in storage and movement of molecules
Golgi Apparatus
Helps package proteins and lipid molecules
Endoplasmic Reticulum
Produce proteins
Rough ER
Rough ER had ribosomes on it, they produce proteins.
Smooth ER
Synthesize lipids and helps in detoxification
Vacuoles
Mitochondira
Turns glucose into ATP
Cytoskeleton
Helps maintain cell shape and stability
DNA
Animal Cells
Centresomes
Lysosomes
Organelle that breaks down things with digestive enzymes.
Plant Cells
Plasmodesmata
Key in movement of molecules between cells.
Large Central Vacuole
Chloroplasts
Cell Wall
Gives structure/support to cell
Subtopic
They both contribute to cell structure: Lipids form the phospholipid bilayer, while carbs contribute to glycoproteins
Types
Complex
Both provide sources of energy
Raises blood glucose levels for longer and produce a more lasting elevation in energy.
Examples: - Starches - Legumes - Whole Grains
Simple
Quick bursts of energy due to the body being able to metabolize quickly
Examples: - Sugars - White bread
Structure
Monosaccharides
Hydrogen
Oxygen
Carbon
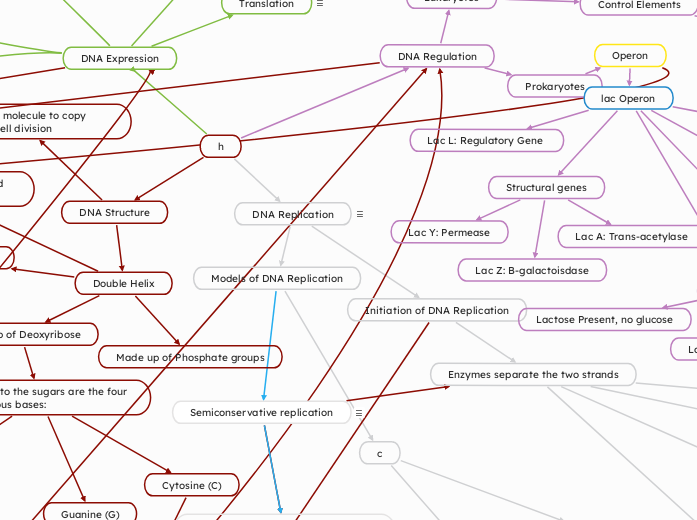
Two strands of the parental molecule separate and function as a template for synthesis of a new, complementary strand
Each parental strand serves as a template for a new complementary strand.
Nucleotides complementary to the parental strand are connected
Form the sugar-phosphate backbones of the new daughter strands
links histones together to form the nucleosome
Transcription
No transcription
Process by which a cell copies its DNA to produce two identical copies
Conservative Replication
Two parental strands reassociate after functioning as templates for new strands
Restore the parental double helix
Dispersive Replication
Each strand of both daughter molecules contains a mixture of old and newly synthesized DNA
Helicase unwinds and separates parental DNA strands
Next, there is a formation of a daughter strand or a new polymer of DNA
Many Okazaki fragments are made at the lagging strand
DNA pol I removes the RNA primer and replaces it with DNA nucleotides
DNA ligase seals gaps
Synthesis of Leading Strand
After RNA primer is made, DNA pol III starts to synthesize the leading strand
Leading strand is elongated continuously as the fork progresses
DNA Polymerases
Two DNA polymerases needed in bacterial replication
DNA Polymerase III
DNA Polymerase I
Need sliding clamp
Converts DNA pol III from being distributive to processive
Need RNA primer to add nucleotides to
Nucleotides added to 3' end of primer
Polymerization occurs in 5' to 3' direction
Add complementary base to daughter strand
Primase synthesizes RNA primers and uses parental DNA as a template
Single-strand binding proteins stabilize unwound parental strands
Topoisomerase breaks, swivels and rejoins parental DNA ahead of replication fork
Relieves the strain caused by unwinding
Translation is the synthesis of a protein from an mRNA template.
Enzymes and Factors
Elongation factors
The process of synthesizing RNA from a DNA template.
Enzymes and Factors:
Prokaryotes: RNA polymerase.
Eukaryotes: RNA polymerase II, general transcription factors, spliceosome (for RNA splicing).
Chargaff's Rule (1950)
Purpose: To analyze the composition of DNA and determine its structural characteristics.
Observations:
The amount of adenine (A) equals the amount of thymine (T), and the amount of cytosine (C) equals the amount of guanine (G): A = T and G = C A = T and G = C
The ratio of purines (A and G) to pyrimidines (T and C) is constant: (A + G) = (T + C) (A + G) = (T + C)
Base composition varies between species, suggesting a role in genetic diversity.
Hershey and Chase Experiment (1952)
Purpose: To confirm that DNA, not protein, is the genetic material.
Method: Used bacteriophages (viruses that infect bacteria) and labeled their components with radioactive isotopes:
³²P: Labeled DNA (phosphorus is present in DNA).
³⁵S: Labeled protein (sulfur is present in proteins but not in DNA).
The phages were allowed to infect bacteria.
After infection: The mixture was agitated in a blender to separate the phage protein coat from bacterial cells. The solution was centrifuged to isolate the bacterial cells.
Results: Radioactive ³²P was found inside the bacterial cells, indicating DNA had entered. Radioactive ³⁵S remained outside in the phage coats, indicating protein did not enter.
Conclusion: DNA is the genetic material responsible for heredity.
Griffith Experiment (1928)
Purpose: To demonstrate the phenomenon of transformation in bacteria.
Method:
Used two strains of Streptococcus pneumoniae:
S strain: Smooth, virulent (caused pneumonia).
R strain: Rough, non-virulent. Griffith injected mice with: Live R strain (non-virulent): Mice lived.
Live S strain (virulent): Mice died.
Heat-killed S strain: Mice lived.
Heat-killed S strain + live R strain: Mice died, and live S strain bacteria were recovered from their blood.
Results: The non-virulent R strain was transformed into the virulent S strain by a "transforming principle" from the heat-killed S cells.
Conclusion: This experiment suggested the existence of a genetic material responsible for transformation, later identified as DNA
Meselson and Stahl Experiment (1958)
Purpose: To determine the mechanism of DNA replication (conservative, semiconservative, or dispersive).
Method: Nitrogen isotopes (¹⁵N, heavy, and ¹⁴N, light) were used to label DNA. Bacteria were grown in ¹⁵N medium to incorporate the heavy isotope into their DNA. They were then shifted to ¹⁴N medium, and DNA was isolated after one and two rounds of replication. The DNA was analyzed using density gradient centrifugation.
Results: First generation (after one replication cycle): All DNA had an intermediate density, ruling out conservative replication. Second generation (after two replication cycles): Half the DNA had intermediate density, and half had light density, confirming semiconservative replication.
Conclusion: DNA replication follows the semiconservative model, where each daughter molecule contains one original strand and one newly synthesized strand.
Introns (Removed)
Exons (Expressed)
Silencers
Promoter
Distal
Bind to specific transcription factors (activators/repressors)
Enhancers
Proximal
Bind to general transcription factors
Nucleosomes
30nm fiber
300nm fiber
Metaphase Chromosome
Specific
Repressors
If there is a high level of transcription, reduces levels
Activators
Increases levels of transcription
General
(Basal/background) low levels of transcription
3' -> 5'
5' -> 3'
Attached to the sugars are the four Nitrogenous bases:
Thymine (T)
Guanine (G)
Cytosine (C)
Adenine (A)
Connected by chemical bonds