Gene expression and G-Protein pathway, I found that the way RNA polymerase 2 binds into the promoter in order to activate gene expression is a lot like how a GTP binds to adenyl cyclase in the G protein pathway, the specific transcription factor, activator in this case, acts like the signal molecule, triggering high level expression.
I find that in gap junctions, molecules are easily permeating through it. We learned that molecules are transported through ion channels, therefore that means that gap junctions must have lots of ion channels as opposed to other junctions.
I find that cell signaling and cell expression have a lot of similarities regarding the way they can be stopped or “turned off”. The cell has a way of regulating itself so that if something goes wrong or if something is unneeded, it can easily make adjustments that can change or modify.
In cell communications, the way a cell signal transduction pathways are very similar to how phosphorylation cascades work. When a cell needs to communicate, it’s interesting how it relays a message whether it's close or far away and how it adapts to the distance.
Another connection I made was between glycolysis and photosynthesis as they are both important processes that are vital to energy production. They are both involved in the creation of ATP, and are both tightly regulated by the cell to balance energy production and demand, with photosynthesis using ATP and NADPH in the Calvin cycle to regulate, and with glycolysis using high levels of ATP or citrate to regulate itself when energy is sufficient.
One connection I made was between Gene expression and regulation and the Meselson-Stahl experiment. This is because in the Meselson and Stahl experiment, researchers demonstrated that DNA is semi-conservative, meaning that each new DNA molecule consists of one old strand and one newly synthesized strand, which is a process that is fundamental for maintaining genetic code during cell division, making sure that the information required for gene expression is accurately passed on to daughter cells.
Another connection I found was through entropy and the breaking of hydrogen bonds. If the hydrogen bonds in nitrogenous bases were to be broken apart, then this would mean that entropy or the amount of disorder would increase in the system. This would increase the entropy because by breaking the hydrogen bonds, they are less structured meaning that they have more freedom. More freedom = more entropy/disorder.
One of the connections I found was through archaea and enzymes, more specifically thermophiles. We know that thermophiles can survive in extreme conditions, like high heat. This has to do with enzymes because usually when enzymes are exposed to high heat, they begin to denature/break down since they can only work at an optimal temperature. There is no change to the enzymes even after being exposed to the conditions thermophiles live in, since thermophiles continue to live/survive. Even if there was change done to the shape of the enzyme, it would continue to work in these conditions as opposed to our enzymes, which would denature if exposed to the same conditions.
DNA structure, replication, expression, and regulation
Found in cells
Membranes, energy, and cell communication
Eukaryotic
Animal Cells
Has a Plasma membrane
Cell membranes are selectively permeable barries
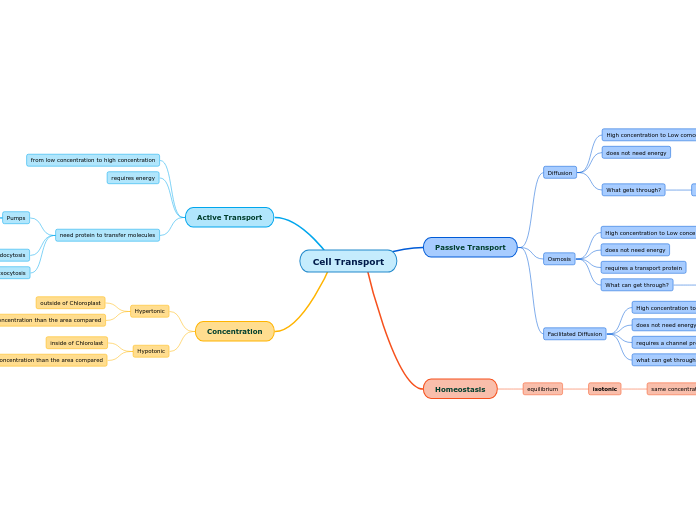
cell transport
active transport
protein pumps
Na+ and K+ pump
establishes proton gradient
2 K+ in
3 Na+ out
ion channel
form pores in cell membranes, allowing ions to pass through
passive transport
diffusion
simple diffusion
small molecules move from high to low concentrations
facilitated diffusion
uses specialized proteins to move ions and molecules across membrane
osmosis
water molecules move with gradient
controls what goes in and out of cell
Has a Golgi Apparatus
Contains Lysosomes
Has a nucleus (nuclear envelope, nucleolus, and chromatin)
Contains Ribosomes
Contains Peroxisomes
Has a Mitochondria
Plant Cells
Has a Golgi apparatus
Contains both smooth and rough ER
Has a nucleus
Has a central vacuole
Has Plasmodesmata
Cell Respiration (Energy Making Process)
In plant
Photorespiration
C4 Plants
Use PEP carboxylase
instead of rubisco cause it
favors CO2 over oxygen
CAM Plants
Photosynthesis during the day
Takes in CO2 at night
in hot and dry conditions the stomata are partly closed due to which CO2 concentration is low in cells. Rubisco favors to bind O2 instead of CO2, so if CO2 concentration is low, Rubisco will bind whatever O2 is present, releasing CO2
Calvin Cycle
Phase 1: Carbon Fixation
3 CO2
Interacts with Rubisco
6 3-Phosphoglycerate
6 1,3-Bisphoglycerate
6 P
6 NADP+
Phase 2: Reduction
6 Glyceraldehyde-3-phosphate
(G3P)
Phase 3: Regeneration of�the CO2 acceptor (RuBP)
3 ATP is used
Ribulose bisphosphate�(RuBP)
1 Glyceraldehyde-3-phosphate
(G3P) leaves the chain to become sugar
6 ATP is released
Electrons are gained from the break down
of H2O
Photosystems
Photosystem 2
P700
Accept light of wavelength 700nm
Photosystem 1
P680
Accept light of wavelength 680nm
Light Reaction
If excess NADPH
Start in PS1
Cyclic flow of electrons is
used to make more ATP, not NADPH
If no excess NADPH
Starts in PS2
Photons go through stroma
Excites electron
Energy gets transfer through chain of stroma with
each excitement of electrons
Energy reaches special pair of chlorophyll
Electron is grabbed by electron acceptor
Transfer down electron transport chain to
PS1 chlorophyll, going through cytochrome complex on the way
1 ATP
Using ATP, H+ is pumped through the thylakoid
spoce against their concentration gradient
Goes through ATP synthase
H+ goes to the stroma, down their concentration gradient
Same reaction happens as in PS2
Electron goes down second electron transport chain
Goes through NADP+ reductase
NADPH
Enter calvin cycle
In animal
Oxidative Phosphorylation
Oxidative Phosphorylation is Chemiosis coupled with ETC
Chemiosis
This happens with all the protons that were
pumped against their concentration gradient
Goes through ATP Synthase
The energy released is used to add a Phosphate to ADP
Electron Transport Chain
This happens to all the electron carriers in the cells
Glycolysis
Starting organic molecule (Glucose)
(C6H12O6)
NAD+ (already in cells)
Redox reaction happens, oxidation of gluclose
(C6H12O6 + 6O2 -> 6 CO2 + 6 H2O + Energy)
Oxygen is reduced
Glucose is oxidized
Glucose
interacts with Hexokinase
Glucose 6 Phosphate
Interact with Phosphoglucoisomerase
Fructo 6 phosphate
1 ATP used
Interact with Phospho-fructokinase
Fructo 1,6-biphosphate
Interact with aldolase
Dihydroxyacetone
phosphate (DHAP)
Glyceraldehyde
3-phosphate (G3P)
Prokaryotic
Is branched into:
Archae
Has ether bond for its Phospholipid
Can survive in extreme conditions
Methanogen can tolerate high acidity
Thermophiles can tolerate high heat
Halophiles can tolerate extreme saline environment
Has branching cell wall
Has Circular DNA
Bacteria
Has Peptidoglycan in cell wall
No membrane bound organelles
Has no circular DNA
No Nucleus
Has plasmid
Has nucleioud
Has fimbria or pili for DNA exchange
Has slime layer or capsule
Has ribosome
Read tRNA and translates it into amino acid chains
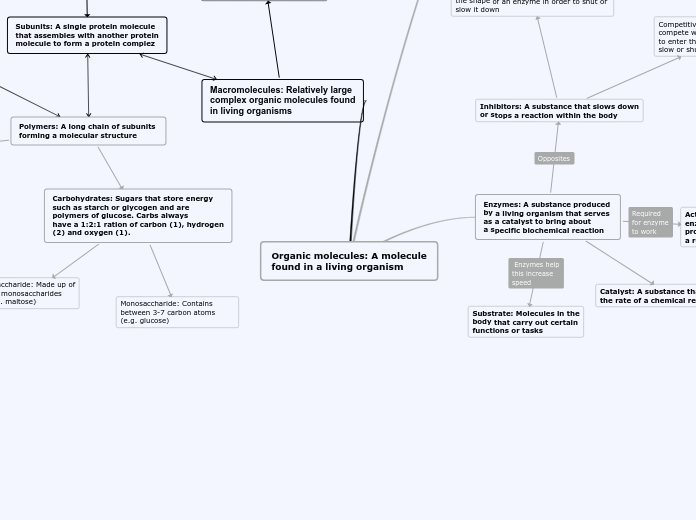
amino acid
comprised of:
H
amino group
carboxyl group
R group
monomer of protein
made of RNA and protein
used for protein syntehsis
Has Flagella for movement (the tail)
Can either be
Facultative Aerobic: can do both
Aerobic: can handle oxygen
Anaerobic: can't handle oxygen
Lipids
HDL vs LDL
low density pipoprotein ' bad cholesterol
sat and trans fat can increase LDL
High density lipoprotein 'good cholesterol'
Steroids
ex: cholesterol
precursor to other steroids
common component of membranes
found in animals
contain 4 fused rings
Phospholipids
form closed bilayers in waater
at 3rd OH another group is attached containing a phosphate group
polar
glycerol is linked to 2 fatty acids
amphipathic
hydrophobic parts gather together in H2O
hydrophilic polar head groups
contains both hydrophilic and hydrophobic parts
Unsaturated Fatty acids
common example: oil
Isomers of unsaturated fatty acids
cis fat
hydrogens on the same side of the carbon chain
the presence of double bonds in cis far causes the molecules to have a kink/slight bemd
trans fat
hydrogens on the opposite side of the carbon chain
do not have hydrogen atoms at every position
one or more double covalent bonds are found within the carbon chain
are liquid at room temperature
come from plant sources
Saturated fatty acids
hydrocarbon chains are tightly packed together by hydrophobic interactions
common example: butter
saturated with hydrogen atoms at every position
no double covalent bonds
solid at room temperature
Commonly found in animal
Major function is energy storage
Comprised of glycerol and 3 fatty acids
Have geometric isomers
Carboxylic acid group at the end of each chain
an ester linkage connects each fatty acid to an OH in glycerol
Formed by dehydration or condensation sysnthesis
Energy transfer
Anabolic Pathways
Pathways consume energy to build larger molecules.
Biosynthetic Pathways
Photosynthesis
Polymerization
Binding
Enzyme-substrate binding
Substrate enters site and changes shape of protein
Enzyme
Catalytic cycle of an enzyme
Enzyme-catalyzed reactions
Rate of reaction increases as substrate concentration increases
Substrates enter and change site
Substrates are not help very strong, there is weak interaction
Active site lowers energy and speeds up reactions
Substrates are converted to products
Products are released and active site is free for new substrates to enter again
pH can affect enzyme activity
pH affects enzyme activity by influencing the enzyme's shape and ability to bind to substrates. If the pH is too far from the enzyme’s optimal range, it can lead to reduced activity aka denaturation
Temperature can affect enzyme activity
Optimal temperatures
Temperature at which cell works just right
To low (temperature) will cause the cell to have a slow reaction
Too high (temperature) will cause for cell to denature
Can be used to speed up chemical reactions
Enzymes achieve this by lowering the activation energy
Thermodynamics
System + Surroundings = Universe
Laws of thermodynamics
Gibbs' free energy
Energy coupler in cells
Coupling
Exergonic + Endergonic
coupling allows cells to carry out complex and energy-demanding tasks by taking advantage of the free energy released by exergonic reactions (ex: ATP hydrolysis).
Transport work
ATP phosphorylates transport proteins when ATP is added to tranport proteins
Causes higher free energy
Unstable
Inorganic Phosphate + ADP + H20 = ATP with 7.3kcal of energy
Chemical reactions are powered by ATP
Can cause lower free energy
Can cause higher free energy
Free-Energy Change
Chemical Reaction
Enzymes can speed up chemical reactions in cells
Hydrolysis
Diffusion
the movement of molecules or ions from an area of higher concentration to an area of lower concentration
Gravitational Motion
Objects moving spontaneously from higher places to lower places
When there less free energy present then it is more stable
Less work capacity
Change in free energy during a chemical reaction
Change in energy can be calculated by doing G final state-G initial state
When more free energy is present then it is less stable
More work capacity
G= H-TS or H=G+TS
Non-spontaneous Process
Endergonic
Non-spontaneous process
Energy is needed
Change in free energy is positive
In equilibrium
Change in free energy is 0
No net changes occurs
Spontaneous Process
Catabolic
Exergonic
Spontaneous process
Energy isn't needed
Change in free energy is negative
2nd law: Every transfer of energy increases entropy in the universe
Entropy
Measure of disorder
1st law: Energy cannot be created or destroyed
Chemical Reactions
Surroundings
Matter in the rest of the universe and outside of the system.
System
Matter with a region of space, can either be an open or closed space.
Closed system: Only energy can come in and out of its surroundings, but not matter
Open system: Energy is free to come in or out in its surroundings
Catabolic Pathways
Pathways release energy by breaking down larger molecules and turning them into simpler, smaller compounds.
Occurs with the net release of energy
Cellular Respiration
DNA is a molecule of heredity in all organisms
Experiments involving DNA
Meselson and Stahl (1957)
Used density gradient centrifugation to track the replication of DNA, grew E. coli in heavy nitrogen and incorporated it into DNA, then centrigued with light nitrogen to receive their results.
Found that DNA was semi-conservative
Griffith (1928)
Injected rats with 4 different types of cells
S-cells: Pathogenic
R-cells: Non-pathogenic
S-cells (heat killed): Non-pathogenic
Mixture of heat killed S-cells and R-cells: Pathogenic
Found that even though cells are heat killed, they can still transfer their DNA.
Hershey and Chase (1953)
Utilized bacteriophage injected with radioactive protein and radioactive phosphorus
After letting the bacteriophage infect, they centrifuged the infected cells.
Radioactive phosphorus was found in the pellet, with the DNA.
They led to the conclusion that DNA is the genetic material
Radioactive protein was found on the outside and not inside the pellet, where all the concentration is at. Radioactive protein was not entering the cells.
DNA Replication
Dispersive
Both strands of daughter molecules contain old and new DNA
Conservative
Parental molecules reassociate
They both act as templates for the new strands, which restores the parental helix
Semi-Conservative
Parental molecules separate
They both become templates for a new and complementary strand
DNA's structure consists of: Phosphate group, nitrogenous bases, and a sugar backbone
Can be found going from 5' to 3'
Nitrogenous bases can be: Adenine, Cytosine, Guanine and Thymine
Chargaff's rule: Amount of Adenine = amount of Thymine, amount of Cytosine = amount of Guanine
Nitrogenous bases are held together by hydrogen bonds
Gene expression and regulation
At what step can the production of a protein be stopped?
Protein can be stopped at any of these steps
Protein processing
protein enters golgi apparatus through cis side
in Golgi, protein is modified with the addition of tags such as phosphate groups
protein is then exited through trans site, in which it has several destinations
Go back to the ER
Go to membrane protein
Gets secreted
Lysosome to be broken down
protein is regulated
Translation
mRNA
Gets attached free smaller subunit ribosome
it slides down and find the start codon (AUG)
tRNA with Met attaches to it
bigger subunit of ribosome sits on top
Translation starts
if signal sequence is reached
Signal Receptor Protein gets attached to it
translation stops
the entire complex enters ER membrane
translation starts
tRNA enters through A side
its Amino acid gets attached to the amino acid of the first tRNA using peptidyl transferase
exits through the E site
this continues until a stop codon is reached
Translation stops
protein is snipped off from tRNA
SRP protein is snipped off
Transportation out of the cytoplasm
Gene production can be stopped at this point
RNA Processing
pre-mRNA
3'
using Poly A polymerase
Attaches a poly A tail to the sequence AAUAA in the 3'
After 3' and 5' end has been modified, mRNA gets modified by spliceosome
alternative splicing
exons are rearranged
some exons are removed
Exons stay
Introns are removed
5'
gets attached to a G group, called a 5' cap
DNA Packaging
Chromatin modification
Nuclesomes
Formation of Nucleosome
Gene production can be stop here
Histone Proteins
H4
H3
H2B
H2A
H1
Linkage protein
DNA
Transcription
Eukaryote
Proximal control element (promoter)
General transcription factors
RNA Polymerase 2 to activate gene expression
Distal Control Element (enhancer)
Prokaryote
Lac I
Constitutive production of repressor
Promoter
RNA Polymerase binds here to activates gene expression
Operator
Gets binded by
Activator
In presence of Glucose
Glucose blocks adenyl cyclase
No cAMP is produced
CAP is not activated
No Activator is made
Activates enhanced level gene expression
Repressor
In presence of lactose
Repressor binds to lactose instead of Operator
Reverts to baseline level or below gene expression
Lac Z
Lac Y
Lac A
Chemical bonds, biological molecules, cell structures, and functions
Cell Communications
Critical players in cell signaling
Phosphatases
enzymes that catalyze the removal of phosphate groups from proteins by hydrolysis
Kinsases
can take a phosphate from ATP and add something to it to activate it
additional/removal of phosphate groups to and from proteins is one way cells regulate protein functions -> can change the shape of a protein, and thus it's function
enzymes that catalyze the transfer of phosphate groups from ATP to proteins
Second messengers
5) cAMP, a second messenger, activates another protein, leading to cellular response
4) Activated adenyl cyclase converts ATP to cAMP
3) Activated G protein.GTP binds to adenyl cyclase. GTP is hydrolized, activating adenyl cyclase
2) Activated GPCR binds to G protein, which is then bound by GTP, activating the G protein
1) 1st messenger binds to GPCR, activating it
cAMP is a second messenger in a G protein signlaing pathway
relay molecules that carry the message from the first messenger (signal) inside the cell
Receptor
Two types of receptors
Intracellular receptors - In cytoplasm and nucleus
Membrane receptors
ex: Ion channel receptor
3) ligand dissociates from receptor, channel closes and ions no longer enter the cell
2) ligand binds to receptor, channel opens, specific ions can flow through the channel and rapidly change the concentration of that particular ion inside the cell, may directly affect the cell
1) channel remains closed until a ligand receptor binds to it
Ex: G protein linked reeceptor (GPCR)
Transmembrane protein - part is inside, part is outside of the cell
receptors in membrane
Signal molecule is hydrophilic
Present in a target cell that receives the signal molecule
Signaling molecule/signal/ligand
Molecule released by a cell which is received by another cell (target cell)
Local signaling (close proximity)
Synaptic signaling
Paracrine signaling
Stages of signaling
Response
Activation of cellular response
Transduction
Phosphorylation cascade
4) protein phosphatases catalyze the removal of the phosphate groups from the proteins, making the proteins inactive again
3) active protein kinase 2 phosphorylates a protein that brings about the cells response to the signal
2) active protein kinase 1 activates protein kinase 2
1) a relay molecule activates protein kinase 1
an example of signal transduction pathway that uses kinases and phosphatases
signal relayed in molecules in a signal transduction pathway
Reception
Signaling molecule binds to receptor
Long distance signaling
ex: Hormonal signaling
If the cell releasing the signal is far from the cell that has the receptor to receive the signaling
Endocrine signaling
Direct contact
These contacts allow molecules to pass from one cell to the other, allowing the recipient to respond
Plasmodesmata within the plant cell
Gap junctions within the animal cell
Proteins gets transported to golgi appartus through vesicle
no atp required
Enzymes that take part in DNA replication
These 2 convert back and forth
before interacting with Isomerase
Isomerase
Converts all to Glyceraldehyde
3-phosphate (G3P)
G3P is oxidized
Energy is released
G3P interact with Triose Phosphate Dehydrogenase, adding a Phosphate to G3P
Forms 2 1,3-Biphosphoglycerate
Interacts with Phosphoglycerokinase
1,3-Biphosphoglycerate gets oxidized with
Phosphoglycerokinase
2 Phosphates groups added to 2 ADP
2 ATP
2 3-Biphosphoglycerate
Interacts with Phospho-
glyceromutase
2 2-Phospho-
glycerate
Enolase, form Double bonds in the substrate through hydrolosis
2 Phosphoenol-
pyruvate (PEP)
Interacts with Pyruvate
kinase and 2 ADP
2 ATPs
2 Pyruvate
If there isn't oxygen
Fermentation
Pyruvate gets reduced
Lactate/Alcohol
The Cori Cycle
If there is oxygen
Pyruvate Oxidation
Starting Molecule (Pyruvate)
Goes through oxidation
Acetyl CoA
Enters citric acid cycle
Citric Acid Cycle
Starting Molecule: Acetyl CoA
Interacts with Oxaloacetate
Citrate
Attaches to an H20
Isocitrate
Gets Oxidized
alpha-ketoglutorate
Gets oxidized
Interacts with CoA-SH
Succinyl CoA
Interacts with succinyl CoA synthetase
CoA-SH
Succinate
Fumarate
Gets Hydrated
Malate
Oxaloacetate
FADH2
ATP
CO2 release
NADH
CO2
2 NADH formed
Altogether, this creates a strand of DNA
Fold together into a condensed chromatin
Fold into an X shaped chromosome
atp required
Gets reduced
Electrons
Complex Q is reduced
Energy released
Used to pump H+ against their concentration
gradient in complex 1,3,4
Interacts with Oxygen
Forms water
NAD+
FAD+
Summary
In order to start replication we need:
Double Stranded DNA
DNA Polymerase
Helps form polymer of DNA and connects the nucleotides in polymer of DNA.
DNA Polymerase I
Found in bacterial replication
DNA Polymerase II
DNA Polymerase III
Lagging Strand
DNA Polymerase I helps remove RNA Primers and replaces them with nucleotides
DNA ligase helps from having the strand fall apart
Leading Strand
RNA Primer made and DNA Polymerase III makes the leading strand
Leading strand is elongated from 5' to 3' direction
RNA Primer
Uses parental strand in order to start replication.
Helicase
Unwinds and separates DNA strands.
Topoisomerase
Unwinds any DNA that may be stuck, makes sure that everything is not tangled.
SSB
Stabilizes the unwound DNA strands.
ORI (Origin of Replication)
This creates a replication bubble/fork
Histone core protein
Types of cells