Regulation of Gene Expression
RNA processing
Proofreading
exonucleolease function
nucleotide excision repair
1. Enzymes detects and
repairs damaged DNA
2. Nuclease enzyme cuts damaged DNA
at two points and removes it
3. Repair synthesis occurs and DNA poly fills missing nucleotides using undamaged template strand
4. DNA ligase seals the end of the new
DNA with the old DNA
Mutations
Silent
no change in amino acid
The original G is replaced with A causing a mutation
Subtopic
There's no change to the amino acid
Missense
3' TACTTCAAACCAATT 5'
5' ATGAAGTTTGGTTAA 3'
5' AUGAAGUUUGGAUAA 3'
Met- Lys- Phe- Ser - Stop
Amino acid changed from Gly to Ser
Frameshift
3' TACTTAGCAAACCGATT 5'
5' ATGAATCGTTTGGCTAA 3'
5' AUG AAU CGU UUG GCU AA 3'
Met- Asn- Arg- Leu- ... 3'
This translation is shifted due to the addition of the two nucleotides. The entire sequence is now read differently.
Insertion or deletion
inserts or deletes one or two nucleotides but never three
Nonsense
change in DNA
3' TACTTCAAACCGATT 5'
5' ATGAAGTTTGGCTAA 3'
3' TACATCAAACCGATT 5'
5' ATGTAGTTTGGCTAA 3'
5' AUGUAGUUUGGcUAA 3'
Met- Stop
The change in DNA resulted in a stop codon so the rest of the mRNA is no translated
5' AUGAAGUUUGGCUAA 3'
Met- Lys- Phe- Gly- Stop
change in amino acid
Translation
Eukaryotes
- Occurs in cytoplasm
- consist of 5' cap (nucleotide Guanine)
- 3' poly A tail (100-250 Adenine nucleotides)
A small ribosomal unit attaches to 5'cap
tRNA carrying amino acid Methionine attaches to start codon AUG
Prokaryotes
Occurs in cytoplasm
A small ribosomal unit attaches to Shine-Delgarno sequence
tRNA carrying amino acid formyl-Methionine (f-Met) attaches to start codon AUG
Large ribosomal unit attaches to mRNA consisting of A site (entry), P site (attaching) and E site (exit)
Another tRNA arrives at the A site carrying another amino acid and attaches to next codon following AUG
The second tRNA moves to P site and the amino acid of the first tRNA attaches while the first tRNA moves to E site
A new tRNA arrives at the A site with and moves to the P site attaching a new amino acid, forming a polypeptide chain
Elongation occurs until a stop codon (UAG, UAA or UGA) is reached
A release factor binds to the stop codon at the A site and acts a cleavage releasing the polypeptide from the last tRNA
The polypeptide released undergoes a modification process in which it is folded into a mature protein structure and translocated via mitochondria, ER lumen, plasma membrane, or lysosome.
The small and large ribosomal unit dismantles and awaits the next translation mRNA sequence
Central Dogma of Biology
DNA
mRNA
Protein
Transcription
Shared Characteristics
RNA transcript is released and polymerase detaches from DNA
Unwinds the DNA and elongates the mRNA (5'-3') - Direction of transcription
Transcription start site - Downstream (+1)
Transcription template strand (3' - 5')
- = upstream, + = downstream
Prokaryote Characteristics
RNA Polymerase
Binds to promoter
Forms final mRNA
Occurs in Cytoplasm
Transcription and Translation are coupled since they both occur in the cytoplasm
Eukaryote Characteristics
Uses RNA Polymerase II
Forms Pre mRNA, snRNA, microRNA
Poly-A Tail
Formed by Poly-A Polymerase
RNA Processing
RNA Splicing
Alternate Splicing
Used to make different proteins
Removal of Introns
Exons are put together forming mRNA
5' Cap
G-P-P-P
TATA box
The TATA box is a DNA sequence (5'-TATAAA-3') within the core promoter region where general transcription factor proteins can bind.
Transcription occurs in the nucleus
Lagging Strand Synthesis
Okazaki fragments
DNA Structure
Fredrick Griffith Experiment
In 1928, he wanted to create a vaccine for pneumonia
R strain and S strain
S strain has a smooth capsule and is pathogenic while R strain was nonpathogenic and had no capsule.
Heat killed S strain with R strain injected rates died.
Something from the S had to be transferred into the R, making the bacteria lethal. Leading to the idea of transformation Bacteria can take up DNA from its environment
Bacteria Transformation
. Something from the S had to be transfered into the R, making the bacteria lethal . Leading to the idea of transformation . Bacteria can take up DNA from its environment
Bacteria taking DNA from environment
R strain injected rates survived
S strain injected rats died
Origin of Replication
E. Coli Bacteria Cell
Circular DNA
1 ORI
Eukaryotic Cell
long DNA molecule
Multiple Replication bubbles
speeds up replication
Helicase
Separates 2 DNA strands to form a replication bubble. It unwinds the double helix at replication forks.
Single Stranded Proteins
Keeps DNA single stranded. It binds and stabilizes a single strand DNA.
Topoisomerase
It relieves overwinding by breaking, swiveling, rejoining DNA
Primase
Synthesize RNA primers on 5' end to 3' end for the Okazaki strand
DNA Polymerase 3
They add complementary bases to DNA and they add nucleotides on the 3' end (5'-> 3'). They need an RNA primer and need a sliding clamp.
DNA Polymerase 1
removes RNA primer and replaces it with the DNA nucleotides
DNA ligase
It seals gaps in nucleotide with phoshodiester linkage. It joines 3' end of the DNA that replaces primer to rest of the leading strand and joins Okazaki fragments of lagging strand.
sliding clamp
Messleson and Stahl Experiment
Proved Semi-conservative Model of Replication
Disproved Conservative and dispersive models.
Bacteria grown in N15
Bacteria moved to N14
centrifuged in CsCl
First Replication
Band in middle
Dense DNA in middle
Second Replication
Band in middle and top
DNA in less dense and dense area
Hallmarks
Speed
E. Coli has 5 billion bases are copied at 2000 nucleotides per second.
Accuracy
Processivity
sliding clamp and polymerases
Hershey and Chase experiment
Bacteriophage
Phages or viruses that infect bacteria. They are made of DNA and proteins.
grown in 35S
Sulfur for proteins because Proteins have sulfur. The protein was found in the supernatant fluid.
blended then centrifuged to see which was radioactive
DNA in 32P was radioactive not protein in 35S
DNA carried genes not protein
grown in 32 P
DNA is grown in Phosphorous because DNA has a phosphate group. DNA was found in the pellet.
Lytic Cycle
The bacteriophages inject DNA to the bacteria so they make bacteriophages. The protein stay outside of the cell.
inject DNA into host
protein stays in cell
host grows bacteriophage parts and assembles in host cell
Cell bursts open
Chargaff's Rule
Number of Adenine = Number of Thymine
Number of Cytosine = Number of Guanine
Watson and Crick Experiments
Double Helix
antiparallel strans
Floating topic
1, 3 Biphosphate glycerate
3 Phosphate glycerate
2 ATP is formed due to the extra phosphate group being transferred to ADP to form ATP
2 Phosphate glycerate
2 phosphoenol-pyruvate (PEP)
2 ATP is formed by the extra phosphate group on PEP being used with 2ADP to from 2 ATP
2 Pyruvates
Energy input needed to start
Metabolism
Enzyme activity
Allosteric Regulation
Can be inhibitor or activator
Can stimulate or inhibit enzyme activity
Binds at one protein site and affects function of other sites
Feedback Inhibition
Initial Substrate
A
B
C
D
End product(inhibits pathway)
Cooperatiivity
one substrate stabilizes enzyme to help subunits lock on enzyme.
Noncompetitive Inhibition
Normal binding
Competitive Inhibition
Thermodynamics
Free Energy Calculations
G(final)- G(inital)
2nd Law
Energy that is transferred and transformed increases the entropy of the universe
1st Law
Energy can be transferred and transformed but no destroyed or created
System
Gibbs Free Energy
ΔG = 0
Endergonic
energy input needed
non-spontaneous
ΔG > 0
Exergonic
energy released
spontaneous reaction
ΔG < 0
△G= △H-T△S
S= entropy (measure of temp)
T= Temperature (STP 273K)
H= enthlapy(potential of system)
H = G + TS
Closed
Open
Surroundings
Universe
Forms of Energy
Potential energy
Electrons on outer shell have the highest PT
chemical energy in food
due to location or position
Stored energy
Kinetic energy
Light energy
Thermal energy
Motion
Metabolic pathways
Anabolic Pathway
Simple molecules
Polymerzation
Biosynthetic Pathways
Photosynthesis
6CO2 + 6H20 + light energy
Simple to complex
6 O2
Glucose
Catabolic Pathway
Complex molecule
Simple molecule
Cellular Respriation
Glucose + 6 O2
Energy into the system
6 H2O
6 Carbon Dioxide
Starting molecule(A)
Adding an enzyme can catalyze a reaction.
B(intermediate)
C(intermediate)
D(end product)
Action Potential In A Neuron
Step 1 - Resting Phase
The activation gates on the Na+ and K+ channels
are closed, and the membrane’s resting potential is maintained.
Step 2 - Depolarization
A stimulus opens the
activation gates on some Na+ channels. Na+
influx through those channels depolarizes the
membrane. If the depolarization reaches the
threshold, it triggers an action potential.
Step 3 - Rising Phase
Depolarization opens the activation
gates on most Na+ channels, while the
K+ channels’ activation gates remain
closed. Na+ influx makes the inside of
the membrane positive with respect
to the outside.
Step 4 - Falling Phase
The inactivation gates on
most Na+ channels close,
blocking Na+ influx. The
activation gates on most
K+ channels open,
permitting K+ efflux
which again makes
the inside of the cell
negative.
Step 5 - Undershoot
Both gates of the Na+ channels
are closed, but the activation gates on some K+
channels are still open. As these gates close on
most K+ channels, and the inactivation gates
open on Na+ channels, the membrane returns to
its resting state.
Cell Membranes and Selectively Permeable Membranes
Transport Across Membranes
Active Transport
Bulk Transport
Receptor-Mediated Endocytosis
Pinocytosis
Phagocytosis
Cotransport
Coupled Transport by a
Membrane Protein
-Occurs when active transport of a solute indirectly drives
transport of other substances
H+/Sucrose Cotransporter (Diffusion of H+ and Active Transport of Sucrose)
Electrogenic Pumps
A transport protein that generates voltage across a
membrane – membrane potential
(-50 to -200 mV)
Help store energy that can be used for cellular work
Proton Pump
Sodium-Potassium Pump
3 Na+ Out / 2 K+ In
Passive Transport
Ion Channels
Ungated
Always Open
Gated
Voltage-Gated
Open and close in response to changes in membrane potential
Ligand-Gated
Open and close when a neurotransmitter binds to channel
Stretch-Gated
Sense-Stretch, Open when membrane is mechanically deformed
Facilitated Diffusion
Carrier Proteins
Channel Proteins
Osmosis
Aquaporins
Hypotonic Solution
Plant Cells - Turgid (Normal)
Animal Cells - Lysed
Tonicity
The ability of a surrounding solution to cause a cell to gain or lose water
Hypertonic Solution
Plant Cells - Plasmolyzed
Animal Cells - Shriveled
Isotonic Solution
Plant Cells - Flaccid
Animal Cells - Normal
Diffusion
Net passive movement of molecules or particles from regions of higher to regions of lower concentration.
Plasma Membrane
Selective Permeability Of Plasma
Membrane
Intermediate Permeability
Large, Uncharged Polar Molecules (Glucose and Sucrose)
Small, Uncharged Polar Molecules (H2O, Glycerol)
High Permeability
Small Nonpolar Molecules (O2, CO2, N2)
Low Permeability
Ions (Cl-, K+, Na+)
Functions Of Membrane Proteins
Transport
Attachment to the cytoskeleton and
extracellular matrix (ECM)
Intercellular Joining
Cell-Cell Recognition
Signal Transduction
Enzymatic Activity
Fluidity Of Plasma Membrane
Viscous
Saturated hydrocarbon tails
Fluid
Unsaturated hydrocarbon
tails with kinks
Phospholipid Bilayer
Hydrophobic tail
Hydrophilic Head
Cellular Respiration
Glucose + Oxygen -> Carbon Dioxide + Water + Energy
C6H1206 + 6O2 -> 6CO2 + 6H2O + ATP
4)Oxidative Phosphorylation
Occurs in mitochondria
OXIDATIVE PHOSPHORYLATION used for ATP production
ETC
ETC = Electron Transport Chain
Electrons move across integral proteins known as Complex I, III and IV.
Complex 1: NADH -> NAD
Oxidation occurs
Complex 2: FADH2 to FAD
Oxidation occurs
CoEnzyme Q10
Mobile carrier of electrons lying on Complex II
Complex 3
Cytochrome C
Complex 4
O2 is oxidized to make H20 with H protons
Chemiosmosis
ATP Synthase
H Protons move down concentratin gradient to make 28-32 ATP
3)Kreb Cylce
Occurs in mitochondria matrix
SUBSTRATE LEVEL OXIDATION for the production of ATP
Acetyl CoA
Citrate
Isocitrate
a-Ketoglurate
Succinyl CoA
Succinate
Fumurate
Malatate
Oxaloacetate
2)Pyruvate Oxidation
Occurs in mitochondria matrix.
Pyruvate + coA
Acetyl CoA
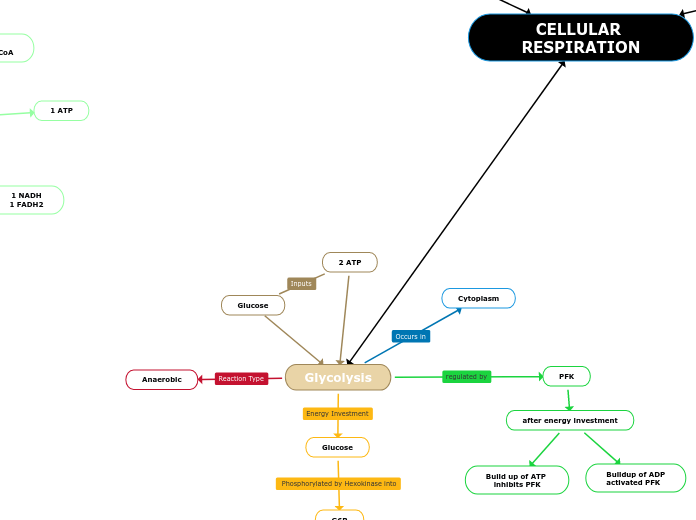
1)Glycolysis
Occurs in cytosol
SUBSTRATE LEVEL OXIDATION to produce ATP.
1)Energy Investment Phases
2 ATP USED
Glucose -> Glucose 6 phosphate
ATP used
Fructose 6 phosphate
Fructose 1, 6 biphosphate
ATP used to convert Fructose 6 phosphate to Fructose 1, 6 biphosphate
DHAP (later forms G3P)
Aldolase
G3P
2)Energy Payoff Phase
4 ATP FORMED
2NAD+ -> 2NADH
Adenylyl Cyclase activated
ATP
Autophosphorylation
6 ATP-> 6 ADP + 6 Pi
Triggers Action Potential
depends on signal/ligand concentration (neurotransmitters)
Second Messenger: cAMP
AMP
Cell Signaling
Physical Contact
Animal Cell: Gap Junction
Plant Cell: Plasmodesmata
Sending Signals using Signals/Ligands and Receptors
Local Signaling
Long Distance Signaling
Receptors
Membrane Receptor
Ion Channel Receptor
Ligand Gated Ion Channel
Tyrosine Kinase Receptor
2 polypeptide dimers (protein kinase)
Tyrosine dimers
G Protein Linked Receptors
Phosphatase for GDP and Pi
GTP binds to GPCR
Signals
Phosphorylation Cascade
Phosphatase
Enzyme catalyzes removable Pi from protein with hydrolysis
Protein Kinase
enzyme catalyzes Pi from ATP to proteins
Activiation of Cellular Response
Amplification effect to be efficient and fast
Intercellular Receptor
Nonpolar signal
Signal Transduction Pathway
Reception
Transduction
ligand or signal is bonded to membrane receptor
PLANT CELL
Plasmodesmata Channels
controls flow of water out of cell
Central Vacuole
cell sap with inorganic ions
stores water and regulates turgor pressure
Cell Wall
made of Cellulose, gives cell structure, keep turgor pressure, control what enters and leaves the cell
Cyanobacterial Prokaryotic
Photosynthetic prokaryotic
- makes their own food with light
Chloroplast
- cell wall is made of cellulose
- performs photosynthesis for energy
- found in the plant domain
Photosynthesis light dependent and Calvin reaction
Mitochondrion Prokaryotic
Oxygen-using nonphotosynthetic prokaryotic
- they are obligate aerobes prokaryotic
Mitochondria
- double membrane
- has its own DNA and ribosomes
- is inherited maternally
- performs ATP synthesis and cellular respiration for energy
ATP Synthesis and Cellular Repiration
EUKARYOTIC CELL
Plasma membrane
permeable lipid bilayer and allows things into and out the cell
has protein and lipids in the bilayer; phospholipids
permeable lipid bilayer and allows things into and out the cell
Endoplasmic Reticulum
Rough ER
bound ribosomes; distribute vesticles; secrete glycoprotein
Smooth Er
synthesize lipids; metabolize carbs
Nucleus
nuclear membrane
nuclear envelope with lamina (α and β) and nuclear pores
nucleolus
ribosomes produced; rRNA transcribed
Chromosome
Chromatin
DNA and histones
PROTEIN SYNTHESIS
RNA produced and leaves nucleous
RNA makes contact with ribosomes (free or bound on Rough ER)
proteins are produced
Vacuoles
Contractile Vacuole
pumps water out of cell
Food vacuole
Golgi Apparatus
packaging proteins
Cis Face
revieves vesicles from ER
Trans Face
releases vesible after altering it
lysosomes
low pH; has enzymes
phagocytosis
autophagy
peroxisomes
metabolized H2O2-> H2O
Oparin’s bubble hypothesis: Simples organic molecules→ complex organic molecules
- Earth had little oxygen, but water vapor and chemicals like carbon dioxide, hydrogen, methane, ammonia, and nitrogen oxides were released by volcanic activity.
- When the volcanoes erupt underwater, the gas bubbles make simple organic molecules that pop when they reach the surface of the water which allows the simple organic molecules to become complex organic molecules which then go back to the sea.
- Early atmosphere was reducing electrons and the volcanic eruption led to organic compounds becoming complex.
- Simples organic molecules→ Complex organic molecules
formation of protein and nucleic acid
Protocell
droplets with lipid bilayer membranes to keep internal chemicals separate for the packaging of proteins and nucleic acids.
Self Replicating RNA
- jumpstarted biological evolution because it carries information and can make a copy of itself allowing for inheritance
- Ribozyme (it’s an dRNA with enzyme function)
Ribozyme
They are RNA with enzyme functions
PROKARYOTIC CELL
- Components of a Prokaryotic Cell:
- Plasma membrane - allows things into and out of cell
- Gas vacuole - to control buoyancy
- Ribosome - protein production
- Flagella - transportation
- Endospore - protects chromosomes from harsh environment
- Inclusion body - storage for glycogen
- Nucleoid - area of genetic material and has chromosome
- Cell wall - peptidoglycan; is modified sugar and amino acid; D enantiomer
- Periplasmic space - oxidation of proteins
- Capsule and slime layer - protects cells from environment
- Fimbriae and Pili - to reproduce
- Nutrition :
- Autotroph
- Photoautotroph: light
- Chemoautotroph: inorganic compounds
- Heterotroph
- Photoheteroph: light
- Chemoheterotroph: organic compounds
- Metabolism of Oxygen :
- Obligate Aerobes: require oxygen for cell respiration
- Obligate Anaerobes: the cell is poisoned by oxygen; fermentation anaerobic respiration
- Facultative Anaerobes: can do both aerobes and anaerobes functions depending on if oxygen is present or not
Nutrition
Heterotroph
Photoheteroph
Chemoheterotroph
organic compounds
Autotroph
Photoautotroph
light
Chemoautotroph
inorganic compounds
Endospores
protects chromosomes from harsh environment
Movement
flagella
cell wall
provides structure to cell and determines what enters or leaves cell
Metabolism of Oxygen
Facultative Anaerobes
- can do both aerobes and anaerobes functions depending on if oxygen is present or not
Obligate Anaerobes
- the cell is poisoned by oxygen; fermentation anaerobic respiration
Obligate Aerobes
require oxygen for cell respiration
Prokaryotic Domains
Bacteria
- they have circular chromosomes
- they can't live in temperatures over 100 degrees Celsius
- they have peptidoglycan in their cell wall
- they don't have a nuclear envelope
- they don't have membrane-bound organelles
Archaea
They are extremophiles, which are archaea that live in extreme environments.
- Archaea lipids are branched and have an ether bond
- they have circular chromosomes
- some can live in temperatures over 100 degrees Celsius
- they have no peptidoglycan in their cell wall
- they don't have a nuclear envelope
- they don't have membrane-bound organelles
Extreme Thermophile
- they live in very hot environments.
Extreme Halophile
live in highly saline environment
Methanogens
- live in swamps and marches
- they produce methane as waste
- they are strict anaerobes ( oxygen is toxic )
ENDOSYMBIOSIS SYSTEM
Host Cell
A cell with a nucleus and Endoplasmic Reticulum.
Miller and Urey Experiment
- Tried to prove Oparin’s hypothesis by replicating early earth atmospheric conditions
- Abiotic synthesis of organic molecules is possible meaning Oparin's bubble hypothesis is possible
- Amino Acids were formed
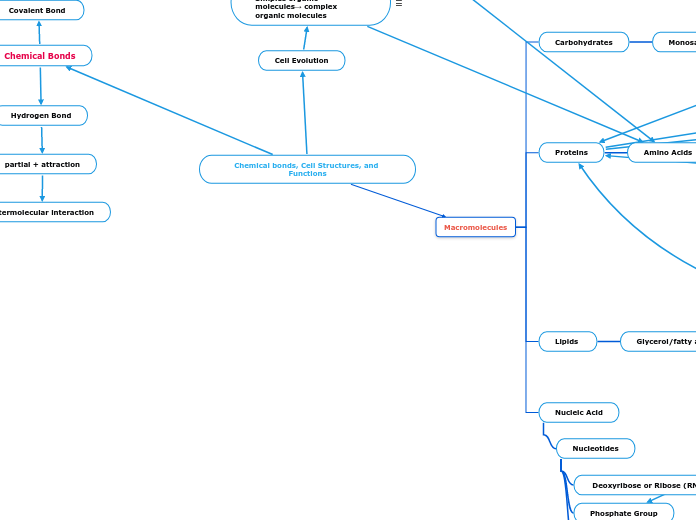
Chemical bonds, Cell Structures, and Functions
Chemical Bonds
Ionic Bond
to obtain a full octet by exchanging e-
Positive
Negative
Hydrogen Bond
partial + attraction
Intermolecular interaction
Covalent Bond
Polar Covalent
e- shared unequally
Hydrophilic
Sharing of e-
Non-polar covalent
Hydrophobic
e- shared equally
Macromolecules
Nucleic Acid
Nucleotides
Nitrogenous Base
Cytosine-Guanine
Adenine-Thymine or Adenine-Uracil (RNA)
Phosphate Group
Deoxyribose or Ribose (RNA) Sugar
Lipids
Glycerol/fatty acids
Trans Fats
Ex: margarine (very processed)
Unsaturated Fats
Ex: butter (solid at Room temperature)
Saturated Fats
Ex: olive oil (liquid at Room temperature)
Proteins
Amino Acids
Side Chain (R Group)
Non-Polar
Ex: CH
Polar
Ex: OH
Basic
Positive Charged R Group
Acidic
Negatively Charged R group
Main Chain
Carboxyl Group
Amino Group
Carbohydrates
Monosaccharides
Polymers - Disaccharides and Polysaccharides
Cell Evolution