Eukaryotic Cells
Bacteria
Gene Regulation
Turns off or reduces the expression of one or more genes by binding to the operator.
Regulate the transcription of genes; Bind to the promotor or enhnacer area of DNA.
Promote gene expression; positive regulation.
Extracellular matrix proteins: collagen
Serum proteins: albumin
Milk proteins: casein
Peptide hormones:Insulin
-Digestive enzymes: Amylase
Examples of secreted proteins
The nuclear envelope is connected to the rough ER which is also continuous with the smooth ER.
Membranes and proteins produced by the ER move via transport vesicles to the Golgi
The Golgi pinches off transport vesicles and other vesicles that give rise to lysosomes, other types of specialized vesicles, and vacuoles.
The lysosome is available for fusion with another vesicle for digestion.
A transport vesicle carries proteins to the plasma membrane for secretion
Endomembrane System
2 ways
-Both processes synthesis is completed on free ribosomes
go through organelles
chloroplasts
peroxisomes
nucleus
1st way: ER (inside of cell "cytoplasm")
Summary:
mRNA--> ribosome-> rough endoplasmic reticulum-->Golgi apparatus--> Lysosome--> Exocytosis
Secretory Pathway: path taken by a protein in a cell on synthesis to modification and then release out of the cell (secretion)
Targeting Proteins to the ER
Polypeptide synthesis begins on free ribosomes in the cytosol
An SRP binds to the signal peptide, halting synthesis momentarily
- SRP is a signal recognition particle and is made of RNA and proteins.
The SRP binds to a receptor protein in the ER membrane, part of a protein complex that forms a pore.
The SRP leaves, and polypeptide synthesis resumes, with simultaneous translocation across the membrane
The signal peptide is cleaved by an enzyme in the receptor protein complex
The rest of the completed polypeptide leaves the ribosome and folds into its final conformation.
Translation
Occurs in the cytoplasm for both eukaryotes and prokaryotes. However, in eukaryotes it is spatially separated from transcription while in prokaryotes it is coupled with it.
Prokaryotes: First tRNA carries formyl methionine; 30s and 50s = 70s ribosome; Has shine dalgarno. Eukaryotes: First tRNA carries methionine; 40s and 60s = 80s ribosome; Binds to 5' end until start codon. Both: Initiator tRNA binds to start codon and large ribosomal subunit joins to form initiation complex.
Polypeptide chains get longer; Ribosomes have 3 binding sites; First tRNA starts at the P site (anticodons of tRNA with codons of mRNA). The A site next to it allows for a tRNA to bind to matching codon forming a peptide bond and then shifting mRNA forward by a codon allowing the empty tRNA to exit through the E site.
Translation comes to an end; Occurs when a stop codon in the mRNA enters the A site. Proteins called release factors recognize stop codons (fit into p site).
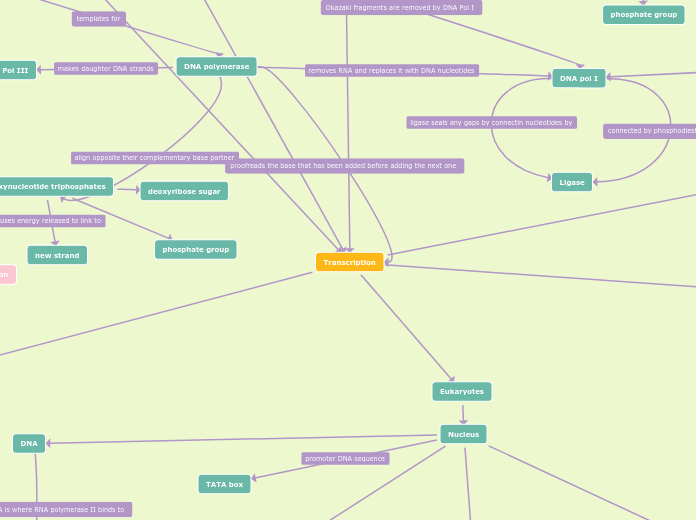
Transcription
Termination
RNA Transcript Releases
mRNA is Released
Poly-A Polymerase Adds 100-200 A's to 3' End Using ATP
Cleavage by Ribonuclease at 3' End
Polymerase Detaches from DNA
Elongation
Moves Downstream Unwinding DNA
Elongate RNA Transcript 5' --> 3'
In Wake DNA Reforms Double Helix
Initation
Promoter Includes Nucleotide Sequence TATA Box
Transcription Factor Must Recognize Before Binding of RNA Polymerase II
Additional Transcription Factors Bind with RNA Polymerase to Form Transcription Initation Complex
About 25 Nucleotides Upstream from Transcription Start Point
RNA Polymerase II - pre-mRNA, snRNA, microRNA
snRNA + Spliceosome + Other Proteins
Splice Together Ends of Introns to Form a Ring and Join Exons Together, Introns Form Ring Separate from mRNA
Cut-Out Intron
Spliceosome Components
RNA Polymerase (RNAP)
RNA Polymerases
Binds to Promoter
DNA Unwinds and RNA Synthesis Begins
Start Point (+1)
Can Be In or Directly After Promoter
Read/Template in 3' --> 5'
Downstream (Right = Positive)
Direction of Transcription
Upstream (Left = Negative)
Eukaryotes
Pre-mRNA
Goes Through RNA Processing
RNA Splicing
Alternate Splicing
Splice Together Exons But Can Leave Out 1 or More to Produce Different Proteins During Translation Through Different Exon Sequences
Introns Cut Out/Exons Spliced Together
Exons = Coding Segment
50-250 Adenine Nucleotides Added to 3' End
Poly-A Tail
Polyadenylation Signal
Protein Coding Segments
Start/Stop Codons
5' Cap
Modified Guanine Nucleotide Added to 5' End
Nuclear Envelope
Prokaryotes
Immediately go to Translation Because of No Nucleus
RNA Synthesis
Lagging Strand
DNA Ligase
Seals Gaps Between Fragments
Okazaki Fragments
Leading Strand
Elongated Continuously
Sliding Clamp
Aids DNA Polymerase III
3 Hydrogen Bonds
2 Hydrogen Bonds
Complementary Base Pair in DNA
Complementary Base Pair in RNA
Complementary Base Pair in DNA/RNA
DNA Structure & Replication
Replication Bubble
Origin of Replication
Replication Fork
Separate Double Strand of DNA
Also has DNA Polymerase I in Bacterial Replication
Remove RNA Nucleotides from Primer and Replaces with DNA Nucleotides at 5' End
DNA Polymerase III
Polymerization 5' --> 3'
Bind to RNA Primer
Bonds Nucleotides to Polymer
Removes 2 Phosphate from Nucleotide
2 Inorganic Phosphate Produced
Forms Phosphate Group/Backbone
Primase
Form Okazaki Fragments of Lagging Strand
Synthesize RNA Primers 5' Leading Strand
SSB
Stabilize Unwound Parental Strands
Topoisomerase
Breaks, Swivels, and Rejoins DNA Ahead of Replication Fork Relieving Strain from Unwinding
Helicase
Unwinds and Separates Parental DNA
Double Stranded
Deoxyribose (DNA)/Ribose (RNA) Sugar
Nitrogenous Base
Cytosine
Uracil (RNA)
Thymine (DNA)
1 Nitrogen Ring
Guanine
Adenine
2 Nitrogen Rings
Phosphate Group
Lactic Acid Fermentation
- Occurs in cytoplasm
- Input: 2 pyruvates, 2H+, 2 NADH
- Output: 2NADH+, 2 lactate
- Net: NAD+
- No ATP is made
Alcohol Fermentation
- Input: 2 pyruvates
- Output: 2 acetaldehyde, carbon dioxide, 2 ethanol
- Net: Ethanol. carbon dioxide and NAD+
- No ATP is made
- Glucose is converted to pyruvate by glycolysis. Pyruvate is converted to ethanol and carbon dioxide
- Occurs in cytoplasm
Fermentation
Water, H20
Energy released
Complexes I,II, and IV
intermembrane space
ATP Synthesis
Proton motive force
Pi
ADP
Chemiosmosis
- Chemiosmosis couples the Electron Transport Chain to ATP synthesis
Oxaloacetate
Citrate
Isocitrate
Redox reaction: Isocitrate is oxidized; NAD+ is reduced
Redox reaction: After CO2 release, the resulting four- carbon molecule is oxidized (reducing NAD+), then made reactive by addition of CoA
There is an addition of a phosphate to Succinyl CoA which causes GTP to be released and bind with ADP which forms Succinate
Redox reaction: Succinate is oxidized; FAD is reduced
Addition of H20 to Fumarate
Malate
Redox reaction: Malate is oxidized, NAD is reduced
Pyruvate oxidized
NAD+ to form NADH
Acetyl Coenzyme A
Citric Acid Cycle/ Krebb's Cycle
- Input: 2 acetyl CoA, 6, NAD+, 2 FAD and ADP
- Output: 2 ATP, 6 NADH, 2 FASH2
- Net: 6 NADH, 2 FADH, 2 ATP
- ATP is made
- Steps 1 and 3 are important
inside mitochondrion
(2) Using energy from this exergonic redox reaction a phosphate group is attached to the oxidized substrate , making a high-energy product
Energy payoff phase
- We are using two molecules of G3P
- We are making more ATP in last 5 steps than used in first 5 steps
(1) G3P is oxidized by the transfer of electrons to NAD+ , forming NADH
Phosphate group is transferred to ADP (substrate-level phosphorylation) in an exergonic reaction. The carbonyl group of G3P has been oxidized to the carboxyl group (-COO-) of an organic acid (3- phosphoglycerate)
Enzyme relocates remaining phosphate group
Enolase causes a double bond to form in the substrate by extracting a water molecule, yielding phosphoenolpyruvate (PEP) , compound with high potential energy
Phosphate group is transferred from PEP to ADP (second example of substrate- level phosphorylation) forming pyruvate
Energy Investment Phase
- First 5 steps don't use ATP (no ATP is made)
Addition of phosphate from ATP to glucose6P using the enzyme of Hexokinase
Glucose 6 phosphate converted to fructose 6-phosphate
Uses enzyme PFK (Phosphofructokinase) to convert fructose6phosphate to fructose1,6 bisPhosphate by transferring a phosphate group from ATP to the opposite end of the sugar, investing a second molecule of ATP
Aldolase cleaves the sugar molecule into two different three-carbon sugars
6 carbon sugar splits into two molecules of 3 carbon each forming DHAP and G3P. Eventually DHAP converts G3P , so at the end we have 2 molecules of G3P from 1 molecule of glucose
Oxidative Phosphorylation
- Through Oxidative Phosphorylation NADH and FADH2 carry electrons down the ETC generating ATP
- Input: NADH and FASH2
- Output: 26-28 ATP
- Net: NAD+, fad, H20
- ATP is made
- Electron chain and chemiosmosis are important
Inner mitochondria membrane
Complexes I,II,III,IV and Q
Glycolysis
- Inputs: Glucose, 2 ATP, 4 ADP, 4P, 2 NAD+, 4e-, 4H+
- Outputs: 2 ADP, 2P, 4 ATP, 2 NADH, 2H+, 2 pyruvates, 2H20
- Net: 2 ATP, 2 pyruvates, 2 NADH
- ATP is made
- Key Steps: 1 and 3
Cytoplasm outside mitochondria
- Electrons extracted from glucose (food) and added to electron carrier, NAD+ and occurs in the cytoplasm outside the mitochondria
Pyruvate Oxidation
- Once pyruvate is made if O2 is available (aerobic condition) it enters mitochondria and is oxidized to form NADH and Acetyl Coenzyme A, which then feeds into the Citric Cycle.
- Input: 2 NAD+, FAD, CoA, Pyruvate
- Output: 2 Acetyl CoA, NADH, fADH2, 2 ATP, CO2
- Net: 2 pyruvate
- ATP is made
- Important steps: 1 and 3
In the mitochondrial matrix
Cellular Respiration and Fermentation
Mitochondria Supply ATP
Cell Signaling
Ligand/Signal Molecule
Epinephrine
Smooth Muscle Cell
Cell Relax
Blood Vessel Dilation
Increased Blood Flow to Skeletal Muscle
Beta Receptor
Liver Cell
Glycogen Breakdown
Glucose Release
Blood Glucose Increase
Tyrosine Kinase Receptors
Forms Dimer
6 ATP Activate Tyrosine Kinase Receptors
Phosphorylated Dimer
Active Relay Protein 2
Cellular Response 2
Active Relay Protein 1
Cellular Response 1
Active Relay Molecule
Active Protein Kinase 1
Active Protein Kinase 2
Phosphorylated Protein
Active Protein Enters Nucleus
Binds to Gene as Transcription Factor
mRNA Transcribed
Leaves Nucleus for Ribosome
Ribosome Creates Protein as Response
G Protein Coupled Receptor
Activated GPCR
Binds to G Protein
Activates G Protein
GTP
Hydrolyzed to GDP
Binds to Adenylyl Cyclase
Active Adenylyl Cyclase
Converts ATP to cAMP (Second Messenger)
Activates Protein
Cellular Response
Intracellular Receptors
Accepts Nonpolar Ligands
Membrane Receptors
Accepts Polar Ligands
Photosynthesis
Stroma
Calvin cycle
Regeneration of the CO2 Acceptor (Phase 3)
Reduction (Phase 2)
G3P (Sugar)
Carbon Fixation (Phase 1)
3 phosphoglycerate
Thylakoid Membrane
Light Reaction
Photosystem I
Cyclic
No NADPH
Noncyclic
NADPH:
Electron Transport Chain
Chemosmosis
ATP
Photosystem II
H20
O2
Floating topic
Peptide Bonds
Electrogenic Pumps
Proton Pump (H+)
Generates Voltage (Membrane Potential)
Store Energy for Cellular Work
Membranes
Plasma Membrane
Phospholipid Bilayer
Selective Permeability
Transmembrane Proteins
Alpha Helix
Cytoplasmic C-Terminus
Extracellular N-Termiinus
Attach to CS and ECM
Cell Recognition
Signal
Enzymatic
Bulk Transport
Endocytic
Receptor-Mediated Endocytosis
Pinocytosis
Phagocytosis
Exocytic
Active Transport
Cotransport
Indirect Transport of Other Molecules
Concentration Gradient
Concentration from Low to High
Requires Energy Input
Sodium/Potassium Pump
Ion Channels
Ungated
Gated
Voltage
Ligand
Stretch
Open Potassium Pump
Hyperpolarization
Open Sodium Pump
Depolarization
Passive Transport
Osmosis
Water from Higher to Lower Concentration
Diffusion
Facilitated
Aided by Proteins
Hydrophobic Outside
Hydrophilc Inside
Channel
Carrier
No Energy Input
Hydrophobic Interactions and van der Waals interactions
Primary Structure
Secondary Structure
Beta Pleated Sheets
Tertiary Structure
Hydrogen, Ionic, Dipole-Dipole
Quarternary Structure
Non-Covalent Bonds Between Hydrophilic and Hydrophobic Surface Units
Alpha Helices
Amino Ends Connect to Carboxyl Ends
SH Forms Disulfide Bonds with other SH R Groups (Only Covalent Bond in R Groups)
Ionic Bond
Hydrophilic (Charged)
Cilia helps for prokaryotic cell movement.
Chromosome
Nucleoid
The nucleoid is within the cytoplasm and contains bacterial DNA. The nucleoid regulates the growth, reproduction, and function of prokaryotic cells. Nucleoid is made of DNA, RNA, and proteins. The nucleoid has an irregular shape.
Plasmid
Plasmids are smaller circles of DNA which is transferrable between cells.
Flagellum
Flagella help bacteria move. Flagella may be scattered over the entire surface or concentrated at one or both ends
Cell Wall
The cell wall is outside of the plasma membrane. The cell wall is made of peptidoglycan which is a polymer of modified sugars crossl
Plasma/ Cell Membrane
The plasma membrane is made of phospholipids and proteins. The plasma membrane controls what enters and leaves the cell, provides protection to the cell, and is also the site of many metabolic reactions (example: cellular respiration and photosynthesis).
Capsules or Slime Layers
Glycocalyx: Capsule or Slime Layers. The cell wall is made of many bacteria and is surrounded by a sticky layer made of polysaccharides or proteins. If it's dense and well defined then it's a capsule and if it diffuses is a slime layer. Both sticky outer layers help bacteria to stick (adhere) to a substrate or other bacteria, Sticky outer layers protect against dehydration and some capsules protect bacteria from attacks by the host immune system.
v
Fimbriae are short-hair-like projections on the surface of bacteria that is used to stick to the substrate or to each other.
Function
Fluidity
Viscous
Fluid
Membrane
Permeability
Attachment to cytoskeleton and the ECM
Cell-Cell Recognition
Enzymatic activity
Intercellular Joining
Signal Transduction
Phospolipids
Amphipathic
Phosphate Groups
Fatty Acids
Microtubules
Microtubules are the thickest of the 3 components which contain tubulin polymers. Microtubules are hollow rods made of a protein called tubulin which there are 2 types (alpha and beta). The microtubules consist of a wall with 13 columns of tubulin molecules. Microtubules grow in length by adding tubulin dimers. They are very dynamic. For example, the dimers can be removed to build a microtubule somewhere else in the cell where its needed. The function of microtubules is the maintenance of cell shape, cell motility, chromosome movement in cell division, and organelle movement.
Microfilaments
Microfilaments (Actin filaments, Actin myosis) are the thinnest component of the cytoskeleton. Microfilaments are at the base of the membrane inside the cell which provides a structural role, changes in cell shape, muscle contraction, cytoplasmic streaming (plant cells), cell motility, and cell division (animal cells). Microfilaments help provide support to the plasma membrane and help bear any tension imposed on the membrane from the outside. The structure of microfilaments is two intertwined strands of actin, thin solid rods mode of protein.
Intermediate Filaments
Intermediate Filaments (fibers with diameters middle range) main function is that helps the maintenance of cell shape, anchorage of the nucleus and certain other organelles; formation of the nucleus lamina. Intermediate filaments are the intermediate of the diameter of microtubules and microfilaments. They are very diverse and made of different proteins such as keratin. The structure is made of fibrous proteins coiled into cables, fibrous proteins supercoiled into thicker cables.
Cytoskeleton
Cytoskeletons' relationship to eukaryotic cells:
The cytoskeleton's relationship to eukaryotic cells is that it maintains the cell's shape and it is the anchorage for many organelles.
Cytoskeleton: reinforces cells' shape, functions in cell movement, and components are made up of proteins.
Endoplasmic Reticulum (ER)
Smooth ER relationship to the Endoplasmic Reticulum (ER):
The Smooth ER's relationship to the endoplasmic reticulum is that its part of the ER and helps with lipid synthesis modification.
Rough ER relationship to the Endoplasmic Reticulum (ER):
Rough ER's relationship to the endoplasmic reticulum is that its part of the ER and helps with protein synthesis.
The Endoplasmic Reticulum (ER) accounts for more than half of the nuclear envelope and help the production and storage of proteins.
Rough ER
The Rough ER has bound ribosomes that separate glycoproteins (proteins covalently bonded to carbohydrates), distribute transport vesicles, and is generally considered the "membrane factory" of the cell. The Rough ER produces proteins, produces enzymes, and surrounds a variety of proteins.
Smooth ER
The smooth ER synthesizes lipids, metabolized carbohydrates, detoxifies drugs and poisons, and stores calcium in muscle cells.
Central Vacuoles
Central vacuoles are present in older plant cells which are eukaryotic cells and they include storage breakdown of waste products, hydrolysis of macromolecules, and enlargement of vacuoles.
Contractile vacuoles
Contractile Vacuoles are found in many freshwater protists, and pump excess water out of cells.
Food vacuoles
Food vacuoles are formed when cells engulf food or other particles.
Vacuoles
Vacuoles Relationship to Golgi Apparatus:
Vacuoles' relationship with the Golgi apparatus is that it is derived from the Golgi and gives rise to the organelle.
Vacuoles' Relationship to the Endoplasmic Reticulum:
Vacuoles' Relationship with the ER is that it is derived from it and membrane/ proteins produced by the ER move via transport vessels.
Vacuoles are large vesicles derived from the Endoplasmic Reticulum and the Golgi Apparatus.
Lysosome Phagocytosis
Lysosome Phagocytosis is when enzymes contain active hydrolytic enzymes which fuse with food vacuoles and hydrolytic enzymes digest the food particles.
Lysosome Autophagy
Lysosome Autophagy is when a damaged organelle becomes surrounded by a membrane so it becomes a vesicle. The lysosome enzymes digest the inner membrane and all the materials of the damaged organelle and release them into the cytosol for reuse.
Lysosomes
Lysosomes' relationship to Golgi Apparatus:
Lysosomes' relationship to Golgi Apparatus is that the Golgi Apparatus receives protein enzymes from the ER, which are packed in a vesicle in the Golgi Apparatus, processed, and then pinched off as a lysosome.
Lysosomes are digestive organelles where macromolecules are hydrolyzed, they are membrane-bound organelles packed with enzymes that are used for hydrolysis to break covalent bonds.
Flagellum
Flagellums' Relationship to the plasma membrane:
The Flagellums' relationship to the plasma membrane is that it also extends from the plasma membrane.
The flagellum is a motility structure present in some animal cells, which are eukaryotic cells, composed of a cluster of microtubules with an extension of the plasma membrane.
Cilia
Cilia's relationship to the plasma membrane :
Cilia's relationship to the plasma membrane is that cilia extend from the plasma membrane.
Cilia help with eukaryotic cells' body movement.
Golgi Apparatus
Golgi Apparatus relationship to the Endoplasmic Reticulum:
The Golgi Apparatus' relationship to the Endoplasmic Reticulum is that it receives proteins and lipids (fats) from the ER.
Golgi Apparatus is active in the synthesis, modification, sorting, and secretion of cell products, stores proteins, and make digestive enzyme for lysosomes.
Plasma membrane
Plasma Membrane relationship to Eukaryotic Cells:
The plasma membrane's relationship to eukaryotic cells is that it provides protection.
The plasma membrane encloses the cell.
Mitochondria
Mitonchondria Relationship to the Eukaryotic cell:
The mitochondria' relationship to the eukaryotic cell is the powerhouse of the cell because they generate energy (ATP).
The Mitochondria are the power house of the cell it produces ATP and is the main site of cellular respiration.
Peroxisomes
Peroxisomes' relationship to the Endoplasmic Reticulum:
Peroxisomes' relationship to the Endoplasmic Reticulum is that peroxisomes emerge from the ER.
Peroxisomes detoxify alcohol in the liver and break down fatty acids and amino acids.
Cytoplasm/ Cytosol
Cytoplasm/ Cytosol Relationship to Eukaryotic Cells:
The cytoplasm is present in all eukaryotic cells that carry out complex metabolic reactions and provides support to the organelle's structures.
The cytoplasm is a gelatin-like substance that provides support for organelles to stay in place and it has structural fibers.
Bound Ribosomes
Bound Ribosomes are attached outside of Rough ER or nuclear envelope, making proteins that are either inserted in the membrane or secreted out of the cell.
Free Ribosomes
Free Ribosomes are located in the cytosol, enzymes that catalyze first steps of sugar breakdown.
Ribosomes
Free Ribosome's relationship to Ribosomes:
Free Ribosome's relationship to ribosomes is that its one type of ribosome based on its location in the cytosol.
Bound relationship to Ribosomes:
Bound Ribosome's relationship to ribosomes is that its the second type of ribosome that is based on being in the Rough ER and the nuclear membrane.
Ribosomes are complexes of ribosomal RNA and proteins, the function in cells is to make proteins. Ribosomes have two locations in cells: cytosol or Rough ER and nuclear envelope.
Nucleus
Connections
Nucleus Relationship to Eukaryotic Cells:
The relationship between the nucleus and eukaryotic cells is that all eukaryotic cells contain a nucleus where they store their DNA and control gene expression.
Nucleus Relationship to the nucleolus, nuclear envelope/membrane, chromatin, centrioles, and nuclear lamina (TEM)
The nucleus connects to the nucleolus, nuclear envelope/ membrane, chromatin, centrioles, and nuclear lamina (TEM) because the nucleus has inside all of these organelles.
Nucleolus Relationship to Nucleus:
The relationship between the nucleolus and nucleus is that the nucleolus is located in the nucleus and assembles ribosomes in the nucleus.
Nuclear Envelope/ Membrane Relationship to Nucleus:
The relationship between the nuclear envelope and the nucleus is that it is a structural framework/ support system for the nucleus.
Nuclear Envelope/ Membrane Relationship to Nuclear Lamina (TEM)
The relationship between the nuclear lamina and the nuclear envelope is that the nuclear lamina provides support to the nuclear membrane so it won't collapse and die.
Chromatin Relationship to Nucleus:
The relationship between chromatin and the nucleus is that the DNA of the nucleus helps make up chromatin which helps make chromosomes when cells divide.
Centrioles' Relationship to Nucleus:
The relationship between centrioles and the nucleus is that the centrioles determine the position of the nucleus.
Centriole's Relationship to Chromatin:
The relationship between centrioles and chromatin both help the process of cell division in the nucleus.
The nucleus stores DNA and controls gene expression.
R Group
Orientation
Interactions
Ionic
Hydrophobic
Hydrogen Bonding
Ion Dipole
Disulfide
Amino Acid
Side Chain
R Groups
Basic
Positive Charge
Acidic
Negative Charge
Polar
OH, NH2, SH, CO
Nonpolar
H, CH, CH2, CH3, H2C, H3C
Main Chain
Carboxyl Group
Ionized
Amino Group
Hydrogen Bonds
RNA Bond
DNA Bond
Nucleoside
Link
Ester Linkage
Beta 1-4 Glycosidic Linkage
Alpha 1-4 Glycosidic Linkage
Alpha 1-6 Glycosidic Linkage
Branch Point
Animal
Plant
Beta Glucose
Alpha Glucose
CELLS
Prokaryotic Cells
Biomolecules
Proteins
Receptor
Hormonal
Contractile/Motor
Structural
Transport
Defensive
Enyzmatic
Nucleic Acids
Nucleotides
ar
Ribose Sugar
Base
Purines
Adenine (A)
Guanine (G)
Pyrimidines
Uracil (C)
r
RNA Only (Replace Thymine)
Cytosine (C)
Thymine (T)
Deoxyribose Sugar
Phosphate
Phosphodiester Bond
Ribonucleic Acid (RNA)
Deoxyribonucleic Acid (DNA)
Complementary Base Pairing (Double Strand)
Allows as Base for mRNA Synthesis
mRNA
Gene Expression (Protein Synthesis)
Directs Own Replication
Lipids
Fat Molecule
Triaglycerol
Fatty Acid (Palmitic Acid)
Unsaturated
Cis
Create C Double Bonds (Kink)
Phospholipids
Hydrophobic Tail
Polar/Hydrophilic Head
Same Side H
Trans
Alternating H
Liquid
Saturated
Solid
Hydrophobic (C-H Bonds. Nonpolar)
Glycerol
Carbohydrates
Polysaccharides
Structure
Chitin
Cellulose
Storage
Dextran
Straight
Branched
Glycogen
Starch
Amylopectin