RNA moves downstream the DNA to continue transcription
Chemical Bonds
Elements
Isotopes
neutrons
Radioactive Isotopes
chemical behavior
# of electrons
valence electrons
valence shells
sharing of electrons b/w atoms in molecules
chemical reactions
products
reactants
Chemical equilibrium
types of chemical bonds
Force that holds 2 or more atoms together
compounds
molecules
organic molecules
monomers
water
universal solvent
hydrophilic
high heat capacity
surface tension
cohesion
adhesion
atoms
ions
protons
neutrons
no charge
+ charge
nucleus
Mass #
Atomic #
polymers
Electrons
negative charge
energy
orbitals
Energy levels
covalent bonds
nonpolar covalent bonds
electrons shared equally & same EN
hydrophobic
Single bond
Double bond
Triple bond
Valence
Electronegativity is when something is an interactive property & the more EN an atom is the more strongly it pulls electrons to itself
polar covalent bonds
electrons not shared equally
hydrophilic
Weak chemical interactions
dipole-dipole
attraction b/w partial + & partial - of 2 molecules
hydrogen bonds
2 polar molecules/bonds
Van der Waals
atoms are close together; very weak
2 nonpolar covalent bonds
Ionic Bonds
Cations
anions
Valence electrons b/w atoms
Strongest bonds in dry compounds!! (known as salts, which may form crystals)
Biological Molecules

Carbohydrates
Monomer: Monosaccharide
Polymer: Disaccharide or Polysaccharide
2 monosaccharide
3 or more monosaccharide
carboxyl group with many hydroxyl groups
Serves as energy source
Storage polysaccharide
Starch (plants)
Amylose (unbranched)
Amylopectin (somewhat branched)
Dextran
Glycogen (extensively branched)
Structural polysaccharide
Cellulose
Chitin
Aldose
Ketose
Lipids
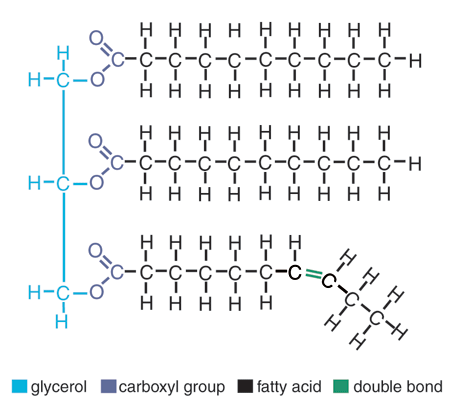
Fat (triacylglycerol/triglyceride)
Fatty acid
Saturated fat
Solid at room temperature
Single bond
Flexible molecule
Unsaturated fat
cis double bond (kink)
Trans fat
formed artificially during hydrogenation
Mostly hydrocarbons
Nonpolar/Hydrophobic
Serves as energy
Phospholipids
Amphipathic
Made up of glycerol joined by 2 fatty acids and phosphate
Hydrophilic head and hydrophobic tail
Steroids

Proteins
Monomer: amino acid
α carbon, amino group, carboxyl group, R group (side chain), hydrogen
Polymer: polypeptide
Primary struture
Amino acid change
Bonds: covalent and peptide
Dictates second and tertiary structure
Secondary structure
α helix
β pleated sheets
Bond: hydrogen bond
Tertiary structure
Interactions between R groups
Hydrophobic interactions
Ionic
Hydrogen
Van der Waals
Covalent (disulfide bridge)
Quaternary structure
Multiple polypeptides
R group interactions
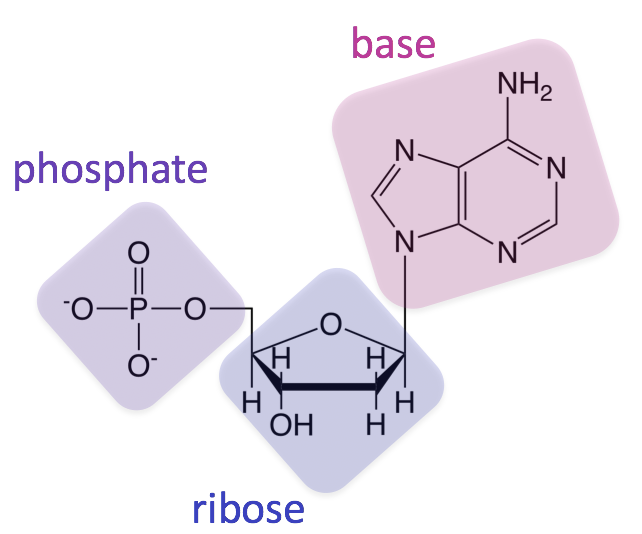
Nucleic Acids
Monomer: nucleotide
DNA
Double strand
Double helix
Genes
Chromosomes
Replication
Daughter strands
Parent strands
Origins of replication
Replication fork
Proteins
Topoimerase
Single-strand binding proteins
DNA polymerases
DNA pol III
Lagging strand
Okazaki fragments
Leading strand
DNA pol I
DNA ligase
Primase
Helicase
Conservative
Antiparallel
Deoxyribose
Complimentary strands
Pentose, phosphate group, phosphate group
RNA
Single strand
Ribose
Nucleoside
Pentose and nitrogenous base
Pyrimidine: six-membered ring
Cytosine (C)
Thymine (T)
Uracil (U)
Fat layers
Fat layers
Purines: six-membered ring plus five-membered ring
Adenine (A)
Guanine (G)
Can function as
catalyst called ribozyme
Self-replicates & stores genetic information about vesicle
Protocell "daughters"
Prokaryotes
Flagella
Plasmids
Nucleoid
Bacteria
Cell wall
Peptidoglycan
Polysaccharides & proteins
Endospores
Archaea
Extremophiles
Extreme thermophiles
Extreme halophiles
Methanogens
Eukaryotes
Animal cells
Extracellular matrix
Cell junctions
Gap junctions
Desmosomes
Tight junctions
Centrosomes
Plant cells
Chloroplasts
Plasmodesmata
Central vacuole
Membrane-bound organelles
Mitochondrion
Cellular Respiration
C6H12O6 + 6O2 -> 6CO2 + 6H2O + Energy (ATP and heat)
Glucose is oxidized
Oxygen is reduced
Glycolysis
Pyruvate Oxidation
Citric Acid Cycle/Krebs Cycle
Oxidative Phosphorylation
Electron transport chain
Proton-motive force
Chemiosmosis
26-28 ATP
6 NADH
2 FADH2
2 ATP
2 ATP
2 NADH
Energy investment stage
Energy payoff stage
4 ATP
Alcoholic fermentation
2 ethanol
2 Acetaldehyde
Lactic acid fermentation
2 lactate
30-32 ATP
Food vacuoles
Peroxisome
Golgi apparatus
Lysosomes
Phagocytosis
Autophagy
Nucleus
Nuclear envelope
Nucleolus
Chromatin
Endoplasmic reticulum
Rough ER
Smooth ER
Cytoskeleton
Intermediate filaments
Maintains cell shape
Formation of nuclear lamina
Anchorage of nucleus
& certain other organelles
Microtubules
Maintains cell shape
Cell motility
Cilia & flagella
Proteins between microtubules
Organelle movement
Kinesin
Vesicle
Motor protein
Vesicle
Chromosome movements
in cell division
Microfilaments
Maintains cell shape
Muscle contraction
Cytoplasmic streaming
Parallel actin filaments
Myosin motor protein
Changes in cell shape
Cell division
Cell motility
Cortex
Inner cytoplasm
Extending pseudopodium
Ribosomes
Metabolism
Facultative anaerobes
Obligate anaerobes
Obligate aerobes
Capsules (Slime layers)
Nutritional Modes
Inorganic chemicals
Chemoautotrophs
Organic compounds
CO2, HCO3-, or
related compound
Light
Photoheterotroph
Photoautotroph
Fimbriae
Pili
newly made RNA elongates
3' end
3' poly-A tail
RNA Splicing
Introns are cut out
Exons are spliced together with a 5'CAP and 3' poly-A tail at it ends
Regular Splicing: exons stay in their natural order
Alternate Splicing: exons can be put together in any order
Essence Hines
ech2748
Functions of molecules in organisms
Biomolecules
proteins
carbohydrates
lipids
nucleic acids
anti body
enzymes
transport/storage
immune system
maintain pH
structural
repairing
collagen
building tissues
messager
Energy production
building macromolecules
sparing proteins and fat
energy storage
steriods
phospholipids
fat layers
fats and oils
unsaturated
Liquid at room temperature
saturated
protecting organelles
provide energy
DNA
RNA
Deoxyribose
store and transports genetic info
Concept Map 2
Molecules involved with Cellular Respiration
CO2 formed
ATP used
Glucose
Oxidative Phosphorylation
Citric Acid Cycle
Mitochondria
Glycolysis
Pyruvate oxidation
2 pyruvate
No ATP used but 1 made
Step 3: Isocitrate is oxidized to form alpha Ketoglutarate
NADH and FADH2 is made
Step 1:Acetyl CoA interact with
Oxaloacetate to form
Citrate
1 Acetyl CoA makes 1 ATP,3 NADH,
3 NADH used
1 FADH2 made
NADH formed
2 Acetyl CoA formed
Pyruvate used
NAD+ used
Water made
ATP made
NADH+ made
ATP is made
2 ATP used
Plasma membrane
Extracellular matrix
Phospholipid bilayer
Phospholipids
Liquid crystaline/fluid phase
Gel phase/rigid
Hydrophobic/hydrocarbon tails
Viscous
Fluid
Hydrophilic head
Cholesterol
Polysaccharide molecule
Proteoglycan molecules
Core protein
Carbohydrates
Extracellular fluid
Proteoglycan complex
Collagen fiber
Fibronectin
Mosaic plasma membrane
Integral proteins
Peripheral proteins
Transmembrane proteins
Functions
Enzymatic activity
Cell-cell recognition
Signal transduction
Attachment to ECM & cytoskeleton
Transport
Passive transport
Osmosis
Water balance
Tonicity
Isotonic
Flaccid
Hypertonic
Plasmolyzed
Shriveled
Hypotonic
Lysed
Turgid
Facilitated diffusion
Diffusion
Carrier proteins
Active transport
Bulk transport
Exocytosis
Endocytosis
Phagocytosis
Pinocytosis
Receptor-mediated
Cotransport
H+/sucrose cotransporter
Channel proteins
Protein pumps
Sodium-Potassium pump
Electrogenic pumps
Proton pump
Aquaporin
Intercellular joining
Selective permeability
Small, nonpolar molecules
Small, uncharged polar molecules
Large, uncharged polar molecules
Ions
membrane
help of molecules from inside
second messenger (help surface receptor)
intracellular
steroid hormone
hormone binds to receptor protein in cytoplasm: activating it
hormone-receptor complex binds to genes
stimulation of gene into mRNA
mRNA translated to specific protein
hormone passes through membrane
CELL COMMUNICATION
Critical players in signaling
ligands
receptors
by releasing a signal
signal receptor proteins
GPCR (transmembrane)
G protein
activation of G protein
Adenylyl Cyclase
cAMP
activates another protein
Phosphorylation Cascade
ligand-gated ion channel
specific ions flow through
cellular response
channel closes
tyrosine kinase receptor
inactive tyrosine dimer
activated dimer
intracellular signal proteins
activated proteins
intracellular signal pathway
stages of signaling
Reception
Response
triggering of phosphorylation cascade
last kinase enters nucleus
transcrip. of specific gene
synthesis of particular protein in cytoplasm
transduction
phosphatase
removal of pi from proteins via hydrolysis
protein kinases
transfer of pi from ATP
by physical contact
signal molecule hydrophilic (1st messenger)
G protein linked receptor
cAMP (second messenger)
Tyrosine Kinase receptor
ion channel receptor
Photosynthesis
6CO2 + 6H2O + light energy -> C6H12O6 + 6O2
CO2 is reduced
Water is oxidized
Light reaction
Photons hit pigment molecules and excite it and causes other pigment molecules around it to get excited to higher energy level
e- transferred from excited P680 to primary e- acceptor
Electric transport chain
Chemiosmosis
ATP
Proton-motive force
Photon hit pigment molecules and excites and excites nearby pigment molecules
e- transferred from P700 to primary e- acceptor
2nd Electron transport chain
NADPH
Carbon Cycle/light dependent
3 CO2 attaches to RuBP with Rubisco to form 6 3-phosphoglycerate
Phase 2: Reduction
6 ADP
6 NADP+ and 6 phosphate groups
6 G3P
Phase 3: Regeneration of CO2 Acceptor (RuBP)
3 ADP
RuBP
C3 plants
Rubisco binds to O2
Photorespiration
C4 plants
CO2 fixed in mesophyll cells and Calvin cycle in bundle-sheath
CAM plants
light reaction at day, Calvin Cycle at night
Water donates 2 e-
6 ATP
6 NADPH
3 ATP
Gene Regulation
Histone acetylation
Promotes transcription by opening up chromatin structure
Condensation of chromatin/reduced transcription
Histone
Histone core
2 H2A, 2 H2B, 2 H3, 2 H4
H1
Prokaryotes
Operons/lac operons
Regulatory genes
lac Z
β-galactosidase
lac Y
Permease
lac a
Transacetylase
Lactose present
Repressor is inactive, RNA polymerase binds and transcription of DNA
Glucose present
No cAMP made to activate CAP so no binding of RNA polymerase and no DNA transcription
No glucose
Production of cAMP that binds to CAP that helps RNA polymerase bind to promoter and transcription of DNA
Lactose absent
Repressor active, RNA Polymerase cannot bind and no trasncription of DNA
Operator
Control access of RNA Polymerase
Activator
Allow RNA polymerase to bind
Transcription of genes
Repressor
Do not allow RNA polymerase to bind
No transcription of genes
Eukaryotes
Specific transcription factors
Proximal control elements
Distal control elements
Enhancer
Allow activators or repressors bind
High transcription
Basal/background expression
Attachment of general transcription factors and RNA Polymerase II
Does not allow for binding of general transcription factors and RNA Polymerase II
Transcription
Initiation
Elongation
Termination
5' CAP
Poly site A
the process of copying a segment of DNA into RNA
Prokaryotes
pre mRNA
microRNA
RNA polymerase
Eukaryotes
RNA polymerase ||
snRNA
5' to 3'/Downstream
RNA synthesis
RNAP
promoter
Translation
mRNA goes to cytoplasm
Ribosomal subunits
protein & RNA
mRNA moves through ribosomes one codon at a time
polypeptide leaves ribosome
P, E, & A site
aminoacyl tRNA synthetase
mRNA has a 5' cap & 3' poly A tail
Elongation
codon recognition
peptide bond formation
translocation
Initiation
modified amino acid f-MET
tRNA base pairs its anti codon with mRNA starting sequence
translation initiation complex (In large ribosomal subunit)
tRNA goes from A site to P site
codons
amino acid codes
termination
release factor
free polypeptide
targeting polypeptide to specific locations
free ribosome in cytosol
SRP binds & attaches
bound ribosome
enzyme signal peptidase
folds into final conformation
ER lumen
golgi
chemical modifications
transported
back to ER
lysosomes
enzymes
plasma membrane
destination in cell