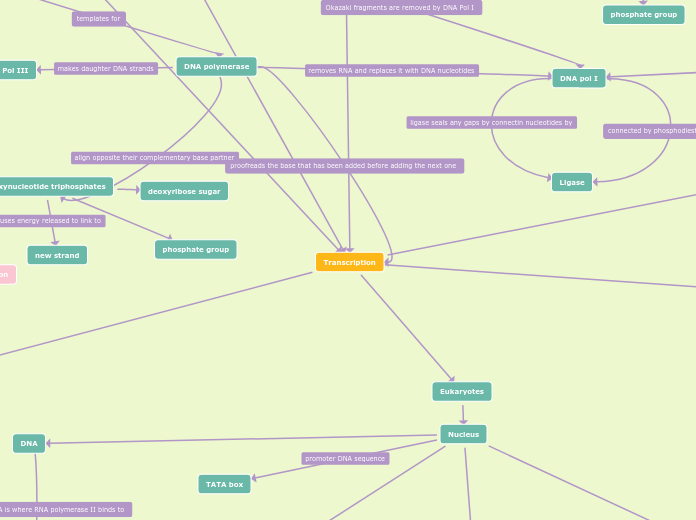
Transcription
Prokaryotes
Cytoplasm
Elongation
Termination
mRNA
Translation
Initiation
F meth= start codon
Eukaryotes
Nucleus
Initiation
meth=start codon
Elongation
Termination
TATA box
DNA
Nucleus
Cell Communication/Signalling
GPCR
First Messenger
G protein
Adenyl Cyclase
cAMP (Secondary messenger)
Protein Kinase A
Cellular Response
enzymes that transfer phosphate group from ATP to proteins
ATP- Adenosine Triphosphate
active= GTP BOUND
INACTIVE= GDP BOUND
Signaling Pathway
signal molecule
receptor protein
relay proteins
Transcription factors
Coding DNA for transcription of a specific gene
Tyrosine Kinase Receptor
Receptor tyrosine kinase proteins
Tyrosines
Tyrosine kinase receptor
Signal Molecule
Kinase
Reception
Transduction
Response
Turn on/off genes
regulate activity of proteins causing/inhibiting gene expression
local
long distance
Feedback regulation
Positive feedback
increased gene expression
Negative feedback
decreased gene expression
Translation
Prokaryotes
Codon Recognition
Eukaryotes
Codon Recognition
Initiation of Translation
Aminoacyl-tRNA synthetases
Mutations
Missense
Nonsense
Frameshift
tRNA
RNA Polymerase II
separates 2 strands of DNA to join complementary RNA
preMRNA
mRNA
Free Ribosomes
enter the endomembrane system by going:
Rough ER
Golgi Apparatus
Lysosomes
out the cell to be used outside
Protien folding
Glycoprotein
RNA Polymerase
Elongation
Termination
Initiation of Translation
Elongation
Termination
Signal peptidase
Cytosol
Mitochondria
Chlorplast
peroxisomes
Nucleus
Biosphere
Communities
populations
Organisms
Organs
Tissues
Cells
Eukaryotic
animal
tight junctions, desmosomes, and gap junctions
cell membrane
made of proteins and lipids
Extracellular matrix
proteins and carbohydrates
Cell communication within tissue and tissue formation
plant
plasmodesmata
cytoplasm
organelles
mitochondria
respiration
chloroplasts
photosynthesis
vacuoles
cell wall
prokaryotic
Eukaryotic cell organelles
plasma membrane
phospholipid bilyar
lipid molecule
fatty acid
saturated
unsaturated
monounsaturated
polyunsaturated
trans-fatty acid
Glycerol backbone
phosphate group
passive transport
Active transport
Osmosis (water)
Facilitated diffusion
Endoplasmic reticulum
smooth ER
rough ER
nucleus
nuclear pores
nuclear envelope
chromatin
nucleoplasm
nucleolus
ribosomes
Free ribosome
ER or Mitochondria, nucleus, peroxisomes, chloroplast, cytoplasm, or out the cell
Membrane proteins
Glycoproteins
Peripheral proteins
Integral Proteins
Transmembrane Protein
Proteins
Golgi apparatus
vesicle
newly forming
secretory
incoming transport
trans face
cis face
cisternae
lumen
Cytoskeleton
filaments
mitochondria
outer membrane
intermembrane space
inter membrane space
inner membrane
DNA
granules
matrix
cristae
ATP synthase particles
lysosomes
membrane
Glycosylated membrane transport proteins
lipid layer
hydrolytic enzyme mixture
peroxisomes
Double membrane
crystalloid core
cytoplasm
cellular respiration
DNA
G Proteins
Cellular response
Ras Protein
Protein Kinases
Transcription
Normal Cell Division
Ras Protein
Protein Kinase
Transcription
increased Cell Division
Upstream
Downstream
Upstream
Downstream
Switch On/off
Actin filaments
Actin
polypeptide subunits
dimers
protofilaments
myosin
microfilaments
microtubules
tubulin
intermediate filaments
DNA Replication
helicase
polynucleotide strand
Lagging strand
Topoisomerase
RNA primer
Single-stranded binding
DNA polymerase
deoxynucleotide triphosphates
phosphate group
deoxyribose sugar
new strand
DNA Pol III
DNA pol I
Ligase
DNA
Prokaryotes
ORI
Eukaryotes
ORI
Bacteria
contains
flagella
Ribosomes
DNA
capsule