Proteins (enzymes) are used to
break down cell waste in lysosomes
glucose reacting with other molecules to form different types of sugars
Cannot occur without
plays a part in
Takes place in
Takes place in
releases
enters A site
translation ends
are moved from one tRNA
to next tRNA
between
forms
As tRNA travels from
A site to P site
Peptidyl transferase
Peptide bonds
Amino acids
Polypeptide chain formed
End codon is reached
Release factor
Complex breaks
apart
tRNA
(w/o amino acid)
Polypeptide chain
Both ribosomal units
attaches corresponding
amino acid to
Aminoacyl tRNA synthetase
begins with
Elongation Begins
first tRNA
starts in
part of
Initiation Complex
initiation complex
complete
Process that creates
binds to
binds to
Contains
tRNA in P site
moves to
here
next tRNA
enters
Sequence
Sequences
Translation
Cytosol
Codons
Sequence of 3 Nucleotides
AUG
Begins Translation
UAA, UAG, UGA
Ends translation
small ribosomal
subunit
tRNA carries
anti codon
Start Codon
(AUG)
Large Ribosomal
subunit joins complex
P site
anti codon pairs with
corresponding codon
E site
(exit site)
tRNA in A site moves to
Transcription
terms
upstream
toward 5' end of DNA
toward 3' end of RNA
downstream
toward 5' end of RNA
toward 3' end of DNA
DNA- dependent RNA polymerase
in 5' to 3' direction
in 3' to 5' direction
DNA polymerase
stop codon
promoter
mRNA
ribosomes
pre-mRNA
introns
spliced out
snRNP's
exons
expressed
a 5' cap made of modified guanine
translation
stability
eukaryotes
comprised of nucleotides made of nitrogenous bases
Metabolism
Anabolism
building up of molecules
requires energy
Catabolism
breaking down of molecules
releases energy
Energy of Life
Chemical Energy
Thermodynamics
entropy always seeks to increase over time
Conservation of Energy
energy cannot be created/destroyed
Photolysis
Two Electrons
Electron Transport Chain
Primary Electron Acceptor
Photosystem I P700
ETC
2 NADH
cell releases
signal molecule
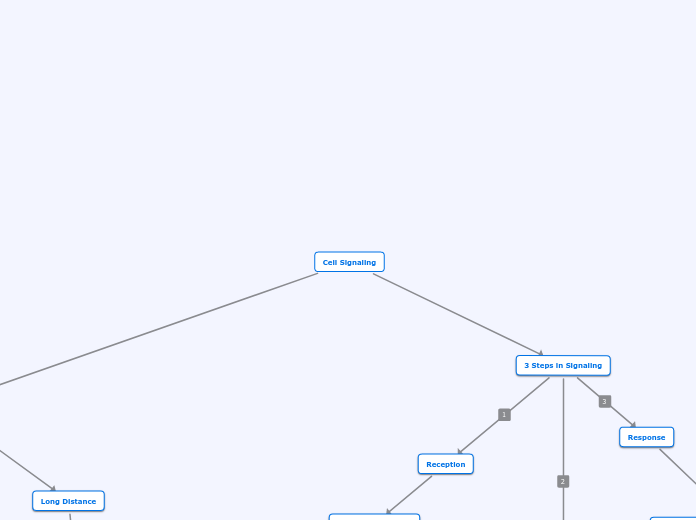
Cell Communication
Physical Contact
Junctions
Desmosomes
Provides a connection
between intermediate
filaments
Somewhere between gap and tight!
Plasmodesmata
Chanel between cells
only found in plant cells
Tight
prevent fluid from moving
across cells.
Gap
Open channel between
cells
Allows free flow of
ions and small molecules
takes place in the
Signaling
Signal Molecule/
ligand
Signal must be received
by a receptor
Signal is non-polar
hydrophobic
Intermembrane
Receptor
1) steroid hormone
passes through the
plasma membrane.
2) hormone binds
with receptor protein
and activates it
3) hormone-receptor complex
enters the nucleus and binds
to specific genes.
bound protein acts
as a transcription factor
4) stimulating the
transcription of
the gene into mRNA
5) mRNA is
translated into a
specific protein.
signal is
polar/hydrophilic
Membrane
Receptor
Tyrosine Kinase
Receptor tyrosine
kinase proteins
(inactive monomers)
(2 of them)
each receptor
has 3 tyrosines
1) 2 Signaling molecules
bind to the 2 binding sites
the monomers combine
to create a dimer
2) 6 ATP is used to
add a Pi to each tyrosine
activating the tyrosine
kinase dimer
2) Inactive relay proteins
attach to the tyrosine dimer
relay proteins become
activated
Triggers Cellular
response
Ion Chanel
signal molecule attaches to the
Ligand-gated ion receptor
channel opens
specific ions are allowed
through the channel into the
cytoplasm
Triggers Cellular
response
When ligand leaves
receptor, channel closes
G Protein
coupled receptor
1) Ligand binds to
extracellular receptor
receptor changes shape
and activates G protein
Activated protein carries GTP molecule
2) activated G protein leaves the receptor, diffuses along the membrane, and then binds to an enzyme
enzyme changes shape
and becomes activated
3) Enzyme takes ATP
and makes cAMP(2nd
messenger)
4) cAMP activates
Protein Kinase A
Triggers cellular
response
Long Distance Signaling/
Hormonal Signaling
Hormone travels through blood steam
to reach target cell
Local Signaling
Paracrine Signaling
Synaptic Signaling
Nerve cell signaling
1) An action
potential arrives,
depolarizing
the presynaptic
membrane.
2) depolarization
opens voltage-gated
channels
Triggering an
influx of Ca2+
3) elevated Ca2+ concentration
causes synaptic vesicles to fuse with
the presynaptic membrane
neurotransmitter is released
into synaptic cleft
4) neurotransmitter binds to ligand-gated
ion channels
Cellular Respiration
dependent on each other as the products of each of these reactions initiate the other reaction
three distinct processes
Citric Acid Cycle
step 1- Acetyl CoA adds its two carbon group to oxoloacetate which produces citrate
step 2- Citrate converts to Isocitrate, with the loss and gain of an H2O molecule
step 3- * Isocitrate is oxidized while NAD+ is reduced. a-Ketoplutarate is made
step 4- CO2 is released from a- ketoglutarate, which causes four-carbon molecules to be oxidized, then CoA is added, which makes it reactive, NADH+ & H+ is formed. CoA-SH is added, creating Succinyl CoA
step 5- A phosphate group is added to Succinyl CoA. GTP is released, which binds with ADP, that results in the formation of ATP. COA-SH is released, and Succinate is the end result.
step 6- Succinate is oxidized, FAD is reduced, FADH2 is formed
step 7- H2O is added to Furmarate, which results in Malate
step 8- Malate is oxidized, NAD+ is added, and it results in the formation of NADH+. The product is Oxaloacetate
Oxidative Phosphorylation
a lot of ATP is formed 26-28 molecules
Chemiosmosis
ATP synthesis
H+ go back down the concentration gradient
Creates ATP
Electron Transport Chain
an electrochemical gradient that leads to the creation of ATP
Complexes I, III, IV
Complex II
FADH2 transfers electrons
their job
pump H+ against the concentration gradient in the intermembrane space
the energy used coms from the energy released from the electrons being transferred down the ETC
are H+ pumps
located in the inter mitochondrial membrane
Pyruvate Oxidation
pyruvate is oxidize, then the electrons are transferred to NAD+ to form NADH
Acetyl Coenzyme A is formed
Glycolysis
Fermentation
Alcohol Fermentation
NADH donates its electrons to a derivative of pyruvate
carboxyl group is removed from pyruvate and released in as carbon dioxide
a two-carbon molecule called acetaldehyde
regenerating NAD+ and forming ethanol
pyruvate decarboxylase and alcohol dehydrogenase
plant tissue, yeast, microorganisms
occurs in production of bread, beer, wine, vinegar
Lactic Acid Fermentation
NADH transfers its electrons directly to pyruvate
glucose is converted to lactate and cellular energy
breaks down glucose into two molecules of pyruvate
cytoplasm
has two phases
Energy Payoff Phase
step 6- 1 G3P is oxidized by the transfer of electrons to NAD+, and forming NADH. Using energy from the redox reaction, a phosphate group, is then attached to the oxidized substrate, making 3-Biphosphoglycerate.
step 7- the phosphate group is transferred to ADP in an exergonic reaction.G3P is then oxidized to the carboxyl group, creating 3-Phosphoglycerate
step 8- Phosphoglyceromutase is used to relocate the remaining phosphate group
step 9- Enolase causes a double bond to form in the substrate by extracting a water molecule, creating phosphoenolypyruvate (PEP)
Energy Investment Phase
step 1- adding a phosphate from ATP to Glucose to form Glucose6p by using the enzyme Hexokinase
step 2- converting Glucose6p into Fructose6p
step 3- * Enzyme PFK is used to convert Fructose6P to Fructose1, 6 Biphosphate
step 4- 6 carbon sugar splits into two molecules of 3 carbon. Each of them forming DHAP and G3P
step 5- conversions between the DHAP
Function:
Photosynthesis
formed when 100 or more monosaccharides are bonded together through glycosidic linkages
Floating topic
amino acid chains and polypeptide backbone held together by
alpha helices, beta pleated sheets
Provides
Can be attached to
Secondary
hydrogen bonding
water properties
universal solvent
water is a polar molecule
water expansion when frozen, denser as liquid than solid
ice has spaced-out lattice structure
ice to float
aquatic life can survive under frozen water
high heat of vaporization
amount of energy to convert 1g or a substance from a liquid to a gas
organisms regulate body temperature by sweating
hydrogen bonds to be broken
high specific heat
amount of heat needed to raise the temperature of 1 gram of a substance 1 degree Celsius
water has highest specific heat of an liquid
hydrogen bonds breaking and reforming
adhesion
water molecules attracted to each other
ex. water move up stem of a plant
cohesion
water molecules attracted to other substances
sharing of H atom
partial positive charges that attract partial negative charges
among H2O molecules
relatively strong
relatively weak
only in
Is attached to the
Surround
Envelope
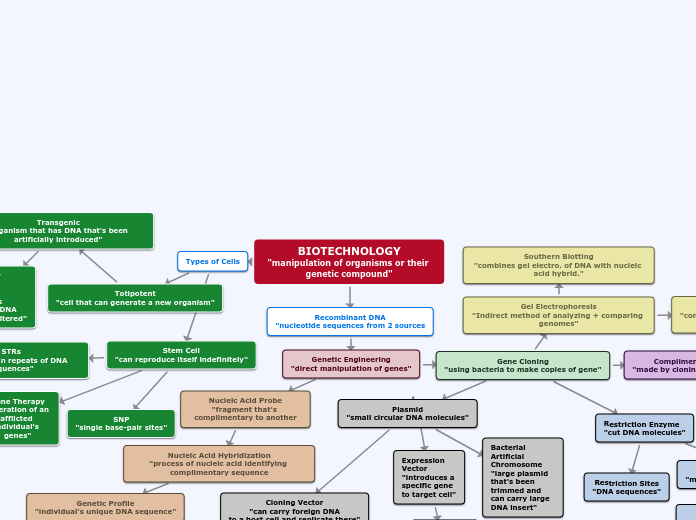
Biological Molecules
Bonds
ionic bonds
covalent
polar
hydrophillic
unequal sharing of electrons
O-H, S-H, N-H, C-O
Include
Nucleic Acids
carry genetic information which is read in cells to make the RNA and proteins by which living things function
Example
DNA, RNA
Nucleotides
nucleic acids
RNA ribonucleic acid)
DNA (deoxyribonucleic acid)
DNA Replication
dna synthesis
helicase
topoisomerase
alters the supercoiling of double-stranded DNA
SSB (single strand binding proteins)
stabilize the un-wound parental strands
primase
DNA polymerase III
sliding clamp protein
converts the DNA polymerase III from being distributive (falling off) to processive (staying on)
DNA Polymerase I
DNA ligase
seals up the sequence of DNA into two continuous double strands
removes the RNA primer and replaces with DNA nucleotides
adds complementary nitrogenous base to daughter strand
leading strand
binds to the leading strand and adds new complementary nucleotide bases (A, C, G and T) to the strand of DNA in the 5’ to 3’ direction
lagging strand
numerous RNA primers, made by the primase enzyme, bind at various points along the lagging strand
chunks of DNA are then added to the lagging strand also in the 5’ to 3’ direction
Okazaki fragments
binds to each strand of DNA at the replication fork and synthesizes a short (3 to 10 base) strand of RNA
primer
provides a strand end for DNA polymerase to add bases to
is an enzyme that catalyzes DNA strand separation
breaking hydrogen bonds between strands
creating a replication fork/bubble for replication to begin
semi-conservative model of replication
The two strands of the parental molecule separate, and each functions as a template for synthesis of a new, complementary strand
Pyrimidines
Thymine (pairs with Adenine in DNA, Uracil (pairs with Adenine in RNA), and Cytosine (pairs with Guanine)
Purines
Adenine (pairs with Thymine in DNA, Uracil in RNA) and Guanine (pairs with Cytosine)
A-T form two hydrogen bonds C-G forms 3 hydrogen bonds
The amount of Adenine equals the amount of Thymine, the amount of Guanine equals the amount of Cytosine
carries genetic information
synthesize proteins
replicates itself
Include 3 Parts
5 Carbon Sugar
Deoxyribose
Ribose
Nitrogen Base
includes phosphodiester bonds
Cytosine
Thymine
Guanine
Adenine
Lipids
serve as structural components of cell membranes, as energy storehouses, and as important signaling molecules
3 Main Groups
Steroids
increase membrane fluidity and serve as signaling molecules
Phospholipid
hydrophillic head, 2 hydrophobic tails
“kinks” in their tails push adjacent phospholipid molecules away, which helps maintain fluidity in the membrane
the structural components of the cell membrane (phospholipid bilayer)
Fat (Triglycerides)
Can Be
Saturated
only single bonds between neighboring carbons in the hydrocarbon chain, saturated with hydrogen
are solid at room temperature
Unsaturated
trans isomer
H atoms on opposite sides of molecule
cis isomer
H atoms on same side of molecule
Presence of double bonds can create cis/trans isomers of fatty acids
stay liquid at room temperature
Proteins
Can Be
Quaternary
several protein chains or subunits into a closely packed arrangement
all prior bonds, van der waals
Tertiary
Subtopic
polar covalent, ionic, hydrophobic interactions, and R-group disulfide bonds
Primary
amino acid chain
peptide bonds
Hemoglobin
Enzymes
Are Chains of
Amino Acids
Known As
Polypeptides
Carbohydrates
serve as fuel and building material
Consisting of 2 or More Sugars
Polysaccharide
Starch
Plants
Glycogen
Formed of Alpha Glucose monomers connected through 1-4 glycosidic linkages
Can be Found Within
Animals
Used For
Storage of Energy
Consisting of 2 Sugars
Disaccharide
formed when a dehydration reaction joins two monosaccharides
Lactose
Sucrose
Consisting of 1 Sugars
Monosaccharide
Examples
Glucose
composed of alpha and beta isomers
Fructose
Can be
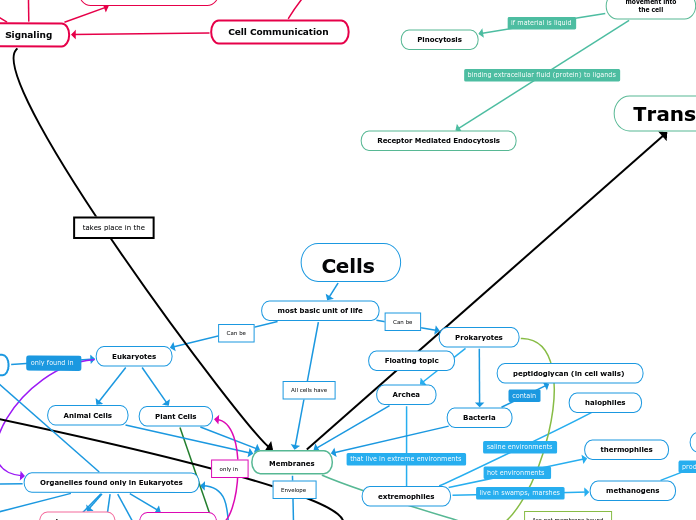
All cells have
Cells
most basic unit of life
Membranes
movement of substances
in and out of the cell
Protein
Integral Membrane
Proteins
Peripheral Membrane
Proteins
Glycoprotein
Carbohydrates
Glycolipid
Lipids
Cholesterol
Phospholipid Bilayer
Amphipathic
Hydrophilic Head
Glycerol
Phosphate Group
Two Hydrophobic Tails
Unsaturated Fatty Acid
double bonds
between carbons
Cis Fatty Acids
H atoms are on
same sides
Trans Fatty Acids
H atoms are opposite
Saturated Fatty Acid
no double bonds
between carbons
Transport
Passive Transport (high to low concentration)
no energy
Osmosis
contractile vacuole
pumping water out
taking water in
water crosses the
the cell membrane
Hypertonic
Plasmolyzed
Shriveled
Isotonic
Flaccid
Normal
Hypotonic
Turgor
Lysed
Facilitated Diffusion
substances cannot cross
membrane on their own
Hydrophobic Molecules
Carrier Protein
shapeshifts to help
different molecules
Hydrophilic Molecules
Channel Protein
uses channel for molecules
and specific solutes
Simple Diffusion
substances easily cross cell membrane
Active Transport (low to high concentration)
energy
large amounts of substances
pass through membrane (vesicles)
Endocytosis
movement into
the cell
Receptor Mediated Endocytosis
Pinocytosis
Phagocytosis
Exocytosis
movement out
of the cell
smalls amounts of substances
pass through proteins (pumps)
Sodium Potassium Pump
conformation/changes in shape
2K+ are brought in against
concentration gradient
3Na+ kicked out against
concentration gradient
Cytoplasm
Organelles
Prokaryotic Organelles
Nucleoid
genetic material, is not membrane bound
Pili
Motion, attaching to objects,
transfer of genetic material
Are not membrane bound
Found in Eukaryotic and Prokaryotic cells
Flagella
Located on the outside of the cell
Allows the cell to move
Ribosomes
Protein Synthesis
Rough ER
Golgi Complex
Glycosylated
Point Mutations
Insertions and Deletions
Frameshift Mutations
Missing/Extra Amino Acid
Extensive Missense
Immediate Nonsense
Base Pair Substitution
Missense
wrong amino acid is produced
Silent
there is no effect on amino acid produced
Nonsense
premature stop codon is present
DNA
RNA
tRNA
Amino Acids
mRNA + tRNA
tRNA on P-site
moves along mRNA making polypeptide chain
Protein is Released
all parts detach for each other
Exit Site
B site
A site
Anticodon
mRNA
Codon
Stop Codon
Start Codon
Uracil
Thymine
Organelles found only in Eukaryotes
Chloroplast
Produces Sugar
ER
Smooth ER
Storage for protein
and lipids
Rough Er
Storage for Protien
Lysosomes
Breaks down cell waste
Vacuoles
Storage
Golgi apparatus
Storage and Transport
Mitochondria
evolved from bacteria
Nucleus
Contains genetic material
Prokaryotes
Archea
extremophiles
methanogens
methane as waste product
thermophiles
halophiles
Bacteria
peptidoglycan (in cell walls)
Eukaryotes
Plant Cells
Photosynthesis
Energy
Light Reactions
Photosystem II P680
CAM Photosynthesis
Malic Acid
Vacuole
takes place at night
H20 loss
C4 Photosynthesis
PEP Carboxylase
Oxaloacetate
Malate
CO2 + Pyruvate
Bundle Sheath Cells
Calvin Cycle
CO2
ADP + P
Glucose
NADP
Stroma
Light-dependent Reactions
Energy from Sunlight
O2
ATP
NADPH
Water
Thylakoid Membrane
Chloroplasts
Chlorophyll
Natural Pigment
Animal Cells