THIS IS APART OF UNIT 2
Subtopic
Link: The primary structure provides the foundation (via peptide bonds), secondary structures emerge from backbone interactions (hydrogen bonds), tertiary structure adds complexity with diverse side-chain interactions ((hydrogen bonds, ionic bonds, hydrophobic interactions, and disulfide bonds ), and quaternary structure assembles multiple polypeptides using the same types of interactions as tertiary structures. These bonds at each level work together to define a protein’s shape and function.
UNIT 2
Also apart of unit 2
UNIT 3
Both Photosynthesis and Cellular Respiration are interconnected since they involve the same components of their formulas, except they are in reverse.
Both are used in:
LINK: Both secretory and cell communication pathways involve signals and enzymes
Unit 3: Point Mutations
Silent
Frameshift
Nonsense
Missense
Unit 3: Signal Pathways: Proteins Transport
Secretory Pathway:
Proteins take this path to synthesis, modification, and then gets released and secreted out of the cell.
The endomembrane system: the plasma membrane, the nuclear envelope, lysosomes, and the endoplasmic reticulum
Protein Modification and Transport
1. The signal sequence peptide gets removed by signal peptidase, and the new peptide is released to the ER lumen.
2. Carbohydrates attach to peptides by multiple enzymes. The resulting protein with the added sugar is called a glycoprotein. (this is called glycosylation)
3. The protein is then packaged into the transport vesicle which gets delivered to the cis face of the golgi apparatus.
4. It is then further modified, and after modification, it is folded into its 3D shape and packaged into another transport vesicle on the trans face of the golgi apparatus.
5. It is delivered to the plasma membrane and gets secreted by the cell.
Secreted Proteins: Examples
ECM Proteins:
-Collagen
Serum Proteins:
-Albumin
Milk Proteins:
-Casein
Peptide Hormones:
-Insulin
Digestive Enzyme:
-Amylase
Targeting Proteins to the ER
1. The polypeptide synthesis begins on a free ribosome
2. The SRP (signal recognition particle) binds to a signal peptide, stopping synthesis briefly.
3. The SRP then binds to a receptor protein located within the ER membrane , which forms a pore.
4. The SRP leaves, synthesis resumes, and translation starts simultaneously across the membrane.
5. The signal peptide is split by an enzyme in the receptor protein complex.
6. The completed polypeptide leaves the ribosomes and folds into the final form.
Unit 3: Translation
tRNA:
A clover shaped RNA that is about 80 nucleotides long, with it's function being to transport Amino acids to mRNA.
Aminoacyl tRNA synthetase:
An enzyme that helps connect Amino acids to the tRNA.
Small Subunit:
Prokaryotes have 30S while Eukaryotes have 40S
Large Subunit:
Prokaryotes have 50S while Eukaryotes have 60S
Ribosome:
Where translation occurs
P Site:
Initiation begins at the P site, where the tRNA attaches to the mRNA in the ribosome and begins the polypeptide chain
A Site:
Where elongation begins, as another tRNA attaches to the mRNA, carrying with it the next amino acids to be added to the polypeptide chain.
Peptidyl Transferase:
An enzyme that makes polypeptide bonds when transferring the amino acids from tRNA to tRNA
E Site:
The exit site for tRNA after it transfers the polypeptide chain to the following tRNA.
Release Factor:
A protein that enters the A site after translation is complete, to break apart the ribosome back into it's subunits
Uses 2 GTP to release newly made protein
LINK: Membrane proteins are synthesized during translation and directed to the ER by a signal sequence recognized by the signal recognition particle (SRP). The ribosome attaches to the ER, and the growing protein is threaded into or across the ER membrane. There, it folds, undergoes modifications, and is packaged into vesicles for transport to its final destination, linking translation to proper membrane protein placement and function..
Unit 3: DNA Replication
LINK: DNA replication depends on the structures and functions of cell membranes, such as compartmentalization, molecular transport, signaling, and energy production. These connections highlight how molecular and cellular systems are integrated to maintain life processes
Replication Fork: Y-shaped structure where replication occurs.
Okazaki Fragments: Short DNA fragments on the lagging strand
Template Strand: The parental strand used to synthesize a complementary strand.
Telomeres: Protective ends of linear chromosomes.
Semi-conservative
The two strands of the parental
molecule separate, and each functions as a template for synthesis of a new, complementary strand.
Conservative
The two parental strands reassociate after acting as
templates for new strands, thus
restoring the parental double helix.
Dispersive
Each strand of both daughter molecules contains a mixture of
old and newly synthesized DNA
Helicase- Unwinds the double helix by breaking hydrogen bonds between bases.
Topoisomerase- Relieves supercoiling ahead of the replication fork.
Single-Strand Binding Proteins (SSBs)- Prevent reannealing of separated strands.
Primase- Synthesizes RNA primers to initiate DNA synthesis.
DNA Polymerase
DNA Polymerase III: Adds nucleotides in the 5′ to 3′ direction.
DNA Polymerase I: Replaces RNA primers with DNA.
Ligase- Seals nicks in the sugar-phosphate backbone, especially on the lagging strand.
Initiation Origin of replication-Specific DNA sequence where replication begins.
-Helicase unwinds the DNA.
-Primase lays down RNA primers
Elongation Leading strand: Synthesized continuously in the 5′ to 3′ direction.
Lagging strand: Synthesized discontinuously as Okazaki fragments
Termination Replication ends when replication forks meet or reach termination sequences.
Ligase joins Okazaki fragments
Unit 3: Gene Expression
Lac Operon: The lac operon is a part of DNA in bacteria (like E. coli) that controls the breakdown of lactose into glucose and galactose.
LINK( to far to physically link): CAP, an activator protein that helps RNA polymerase bind to the promoter in the lac operon, requires cAMP to become active. Adenyl cyclase, which can be influenced by the G-protein pathway, produces cAMP from ATP. If no cAMP is made with the help of the activated Adenyl Cyclase enzyme, CAP cannot bind to the DNA, and RNA polymerase cannot efficiently bind to the promoter, preventing gene expression.
Gene Expression: For the genes in the lac operon to be expressed (used to make proteins), the RNA polymerase needs to bind to the promoter region.
CAP (Catabolite Activator Protein): To help RNA polymerase bind to the promoter, CAP is needed. CAP is an activator protein.
cAMP (Cyclic AMP): For CAP to work, it needs to be activated by cAMP, which comes from the G-protein pathway. When glucose is low, cAMP levels are high.
G-Protein Pathway: The G-protein pathway helps produce cAMP, which activates CAP.
Binding: Once CAP is activated by cAMP, it binds to the lac operon promoter and helps RNA polymerase attach, allowing transcription (gene expression) to happen.
No cAMP: If there is no cAMP (when glucose is high), CAP cannot bind, and RNA polymerase cannot efficiently bind to the promoter, so gene expression doesn’t occur.
Transcription
The creation of mRNA from DNA genes to further go onto be a protein in translation
Eukaryotic Transcription
The Nucleus of a Eukaryotic cell
Several transcription Factors
TATA box ( a nucleotide sequence containing TATA) on the promoter if the DNA
RNA Polymerase II can bind in the correct position and orientation
More transcription factors bind to the DNA along with RNA Polymerase II to form the transcription initiation complex
RNA Polymerase II unwinds the DNA double helix
RNA synthesis begins at the start point on the template strand, adding nucleotides complimentary to the template DNA strand.
RNA Polymerase II moving along the DNA template strand in 3' to 5' direction
RNA polymerase II creates a complimentary RNA strand from 5' to 3', adding nucleotides that match the DNA template strand
The pre-RNA molecule crafted to go through modifications like 5' capping's, splicing, and 3; polyadenylation ( AAA ends) as its being transcribed.
RNA polymerase II reaching the polyadenylation signal (AAUAAA, a code all pre-RNA's have)
the pre-RNA is cleaved downstream(right) of the signal
A poly-A-tail is added to the 3' end of the RNA
RNA Polymerase dissociates from DNA, completing transcription
Prokaryotic Transciption
The Cytoplasm of a Prokaryotic cell
Enzyme RNA Polymerase
the promoter
the DNA strands starts unwinding
RNA polymerase
RNA synthesis at the start point on the templet strand
The RNA polymerase moves
downstream and it..
starts unwinding the DNA and elongating the RNA transcript from 5' to 3'
the DNA strands re-form a double helix
The RNA transcript is released
RNA polymerase detaches from the DNA
a completed full RNA transcript strand from 5' to 3'
At the end of transcription in both eukaryotic and prokaryotic cells, a mRNA strand is made to be translated into a protein during translation
Enzyme active site ( a spacious pocket for bindng)
Enzyme-Substrate complex that weakens the substrates bonds for reactions to occur easier
Every energy transfer or transformation increases the entropy or disorder of the universe!
Makes ATP as energy is needed to make ADP into ATP with the addition of a phosphate group
Exergonic reactions as they are th only reaction that allows cells to do work. REACTIONS NEED TO END UP NEGATIVE
Second Law: (Principle of entropy increase)
Example: Food allows animals to run and release heat which increases disorder/entropy. There would be more order than normal but there is still order. For Biology, we need order.
First Law:
(Principle of conservation of energy)
Example: Consuming food, there is stored energy in the bonds we are eating! potential->kinetic
Energy can be transformed and transferred but cannot be created or destroyed.
ATP; the energy coupler or currency of the cell.
3 phosphates with negative charges
hydrolysis occurs on inorganic phophate
Heat is released so high free energy is left
ATP acts as a Endergonic and exergonic coupler
Unit 2: Cell Membranes
Types of Transport
Passive Transport- Diffusion, Osmosis (water movement),
Facilitated diffusion
Active Transport- requires ATP, pumps (soidum potassium pump)
Bulk Transport- Endocytosis
Phagocytosis (cell eating)
Pinocytosis (cell drinking)
Receptor-mediated endocytosis
Exocytosis
Functions of Selective Permeability
maintains internal environment
Receptor proteins bind signaling molecules
Barrier against harmful substances
Cell Membrane Structure
Tight Junction
Gap Junction
Cholesterol- Provides membrane fluidity and stability
Carbohydrates -help with cell recognition and protection, which are important for selective permeability of the cell membrane
Channel Protein- facilitate the transport of substances across a cell membrane through a process called facilitated diffusion. This process moves molecules from high to low concentration without using energy
Carrier Proteins- transmembrane proteins that move molecules and ions across cell membranes. They are responsible for transporting small molecules from areas of low concentration to areas of high concentration, against a biochemical gradient
Receptor Proteins- a special class of proteins that function by binding a specific ligand molecule. When a ligand binds to its receptor, the receptor can change conformation, transmitting a signal into the cell.
Hydrophilic- water loving heads
Hydrophobic- water repelling tails
Unit 2: Cellular Respiration
Oxidative Phosphorylation
Electrons are removed from NADPH that was formed in previous processes, and put into the electron transport chain(ETC)
Complex 1 -> Complex Q -> Complex 3 -> Cyt C -> Complex 4
The movement of the electron releases energy that pumps protons against its concentration gradient
Chemiosmosis:
Protons are pumped back down their concentration gradient and the energy associated with this movement is used to add a phosphate group to ADP, forming ATP
Citric Acid Cycle
Step 1:
Acetyl coenzyme A enters the citric acid cycle and interacts with oxaloacetate to form citrate
Step 3:
Isocitrate is oxidized and NAD+ is turned into NADH, while isocitrate is processed into ketoglutarate
Energy Production:
3 NADH, 1 ATP, and 1 FADH2
Gylcolysis
Step 1:
A phosphorus from ATP is added to glucose with the help of Hexokinase
to form glucose6P
Step 2:
Glucose6P is turned into fructose6P with the help of phosphogluco-isomerase
Step 3:
Fructose6P is converted into fructose1,6 bisphosphate by using a phosphate from an ATP and the help of phosphofructokinase
Steps 4/5:
The enzyme aldose splits fructose1,6 bisphosphate into glyceraldehyde 3-phosphate (G3P) and DHAP and the molecules are differentiated
Step 6:
G3P is oxidized and an electron is removed from the G3P and given to NAD+ to form NADH, while G3P becomes a byproduct that is given a phosphate group
Step 7:
The byproduct from the previous reaction then gives ADP one of its 2 phosphate groups to form ATP and form another byproduct
Steps 8/9:
An enzyme rearranges the byproduct to make another byproduct which is then processed by enolase to form phosphoenol-pyruvate(PEP)
Step 10:
The phosphate group from PEP is transferred to ADP to make ATP and PEP is turned into pyruvate
Pyruvate Oxidation
Step 1:
Pyruvate is taken from the glycolysis process and moved to the mitochondria and oxidized, giving electrons to to NAD+ to make NADH (requires O2)
Acetyl Coenzyme A:
A byproduct of pyruvate losing an electron, but necessary for the citric acid cycle
Unit 2: Laws of Thermodynamics
GIBBS FREE ENERGY(G): A thermodynamic property that is used to predict the spontaneity of a process based on the principles of the second law.
Free energy Change (ΔG ):
The difference between the free energy of the final state and the free energy of the initial state.
ΔG = G ( final state) - G ( initial state)
Non-Spontaneous: When cell processes need energy to start a process in the cell it is non-spontaneous and ( ΔG < 0)
Endergonic Reaction: Since ΔG is positive (Products>Reactants), it needs to absorb energy to continue the cellular process and bond breakage.
Change = ΔG = ΔH - TΔS ΔH = Enthalpy (total potential of a system)
ΔS = Entropy(measure of temperature)
Equilibrium: ΔG=0, no net change
Spontaneous Reactions: For cell processes to occur without additional energy and its initial state is more than the final state (ΔG > 0 )
Exergonic Reaction: Since ΔG is negative (Reactants>Products) is does not need energy to occur.
Unit 2: Cell Communication
Junctions
Plasmodesmata
The plant versions of Gap junctions, only present in plant cells
Tight Junctions
Junctions that try to prevent leakage and create a semi-permeable membrane which are more strict and selective on which ions get to pass
Desmosomes
Junctions that provide adhesion between cells and allow for more larger ions and molecules to pass through, is an intermediate between gap and tight junctions
Gap Junctions
Junctions on the cell membranes that allow cells to connect to each other and have molecules and substances to go in and out of the cells, resulting in the direct diffusion of ions
Critical Players:
Receptor
The receptor is present in the target cells that is receiving the signal
Intracellular receptor
Membrane Receptor
Signaling Molecule/Ligand
The molecule that is released by the cell which is usually received by another cell. Kickstarts the G--protein pathway.
In G -protein pathway, it Usually binds to a receptor protein on a cell membrane
This process of activating kinases and other proteins is called signal transduction, and it leads to s specific cellular response, gene activation, secretion of substances, or changes in the cell's behavior
LINK( too far to physically link): In the G-protein signal pathway, the signal is passed through a series of molecules inside the cell(signal transduction), eventually leading to a cellular response. This response might involve creating or activating a specific transcription factor. The transcription factor can act as an activator or repressor in eukaryotic cells, influencing transcription by either helping RNA polymerase II bind to the promoter or preventing it from accessing the gene to initiate transcription.
LINK( to far to physically link): CAP, an activator protein that helps RNA polymerase bind to the promoter in the lac operon, requires cAMP to become active. Adenyl cyclase, which can be influenced by the G-protein pathway, produces cAMP from ATP. If no cAMP is made with the help of the activated Adenyl Cyclase enzyme, CAP cannot bind to the DNA, and RNA polymerase cannot efficiently bind to the promoter, preventing gene expression.
cAMP activates kinase enzymes ( like protein kinase A), which add phosphate groups to other proteins inside the cell.
cAMP acts as a message carrier inside of the cell, passing on the signal from the outside
Adenyl Cyclase uses ATP (energy molecule) to make cAMP (cyclic AMP), which is an important secondary messenger.
The activated G-Protein moves along the membrane and turns on an enzyme called adenyl cyclase
The G-protein is a switch. When its inactive, it has GDP attached. Once the receptor is activated, the G-protein gets a boost, and GDP is replaced by GTP, which turns on the G protein
When ligand binds, it activates the receptor protein on the surface, which is connected to a G-protein inside of the cell.
Types of signaling
Long distance signaling
Hormonal signaling
Local Signaling
Synaptic signaling
Paracrine signaling
Enzymes
Enzymes are macromolecules that act as catalysts in chemical reactions in order to speed up reaction rate by lowering activation energy
Substrates
Reactant that enzyme binds to
Energy Payoff Phase:
4 ATP and 2 NADH made
Energy Investment Phase:
2 ATP is used during the processes
Anabolic Pathways
simpler molecules into complex molecules
Ex. Photosynthesis
6CO2 +6H20 + light → C6H1206 + 6O2
Need light energy to create sugar and oxygen
Catabolic pathways
complex molecules into simpler ones
Ex. Cell respiration
C6H1206 + 6O2→6CO2 +6H20 + ENERGY
Feedback Inhibition
prevents a cell from wasting chemical resources by synthesizing more product than is needed.
the product acts as a noncompetitive inhibitor and inhibits the first enzyme of the pathway, causing the pathway to shut down and restrict the creation of more product.
Cooperativity
substrates to control the functionality of an enzymes. The binding of one substrate can active other enzymes subunits into their active forms
Allosteric regulation
enzymes and proteins with quaternary structures
active to inactive depending on the reactions needs
1,4 Glycosidic Linkage:
A linkage between the first
Carbon group of a monosaccharide
and the fourth Carbon group of another monosaccharide
Starch:
Used for storage and
consists of Amylopectin, which creates branches, as well as Amylose which does not branch out
Inhibitors
Non-competitive
binds to the enzyme away from the active site , altering the shape of the enzyme so that even though the substrate can still bind, the active site functions much less effectively, if at all.
Competitive
mimics and competes with the substrate to try and bind with the active site. Overall prevents the substrate from binding.
Enzymes
Ex. sucrose
Unit 1: Biomolecules: Nucleic Acids + Lipids
Lipids
Large biomolecules that include fats, phospholipids, and steroids
Phospholipid
made of glycerol with two fatty acids and a phosphate group; they can form bilayers and function as a membrane
Fat
triglyceride: 3 fatty acids that are linked to a glycerol molecule, fatty acids have long carbon skeletons with a functional group and non-polar C-H bonds in the hydrocarbon chains
Steroid
contains carbon skeleton with four fused rings containing a variety of chemical groups attached
Nucleic Acids
Ribonucleic Acid (RNA)
Single stranded polynucleotide where each nucleotide monomer with a ribose sugar and nitrogenous base
Nitrogenous bases include:
-Adenine (A)
-Guanine (G)
-Cytosine (C)
-Uracil (U)
Pyrimidines:
-one six-membered ring of carbon and nitrogen atom
Members include:
-Cytosine
-Thymine
-Uracil
Deoxyribonucleic Acid (DNA)
Genetic material inherited from parents. Each chromosome from the long DNA molecule contains several hundreds of genes
DNA helps provide instructions for the cell to develop and reproduce by providing the genetic material for all of the proteins that the cell may need.
Double stranded helix where each polynucleotide strand has monomers with deoxyribose sugar and nitrogenous base
Nitrogenous bases include:
-Adenine (A)
-Guanine (G)
-Cytosine (C)
-Thymine (T)
Purines:
-a six-membered ring fused to a five-membered ring
Members include:
-Adenine
-Guanine
Unit 1: Bio-molecules: Carbohydrates + Proteins
Carbohydrates
Cellulose:
Found in plants exclusively
and provides structure, does not branch at all
Beta glucose:
The OH on the first Carbon group
is above the center of the molecule
Glycogen:
Used for storage and
branches extensively
Proteins
Quaternary level: assembles multiple polypeptides using the same types of interactions as tertiary structures.
Tertiary level: adds complexity with diverse side-chain( R-chain) interactions ( hydrogen bonds, ionic bonds, hydrophobic interactions, and disulfide bonds.
Secondary Level: emerge from backbone interactions (hydrogen bonds)
Primary Level: provides foundation (Via peptide bonds)
Unit 1:
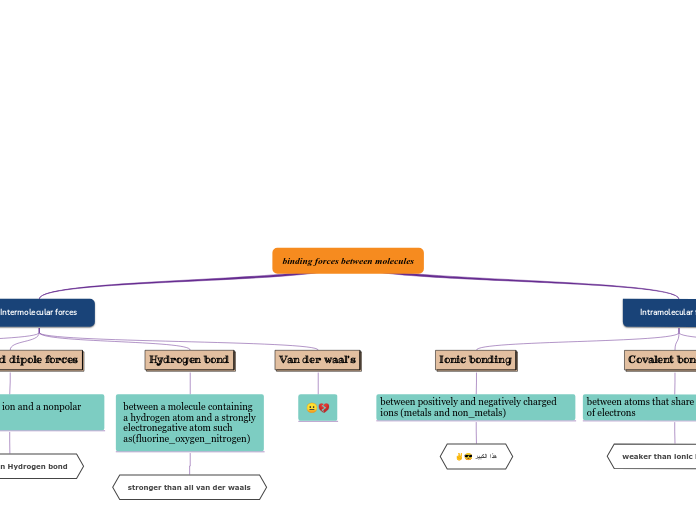
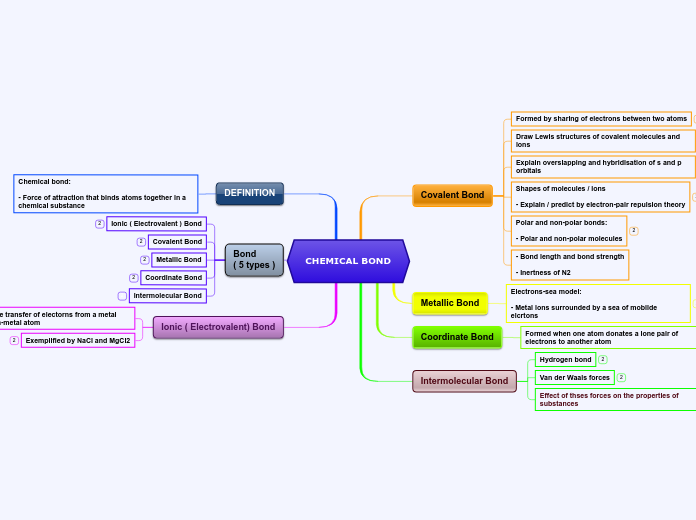
Intramolecular Bonds
(Bonds in-between atoms/element)
(Ex. O-H in H2O)
Covalent
(Sharing of electrons)
Polar
Occurs when there is an unequal sharing of electrons between atoms, within a molecule.
They are still willing to share though!
Example: C-O( 1.0 EN difference), O-H(1.4 EN difference), N-H (0.9 EN difference).
Non-polar
Occurs when electrons are shared equally between atoms, within a molecule.
Example: C-H(Hydrocarbon, 0.4 EN difference), CO2(linear molecular shape).
Ionic
(The attraction of opposite charges, cations and anions)
Occurs when electrons are taken rather than shared between atoms, within a molecule.
Example: Na+ and Cl- make NaCl (table salt)
- The 1 electron in Na+ valence shell if taken by the Cl- to fill its octet.
Intermolecular Bonds
(Ex. Bond between two molecules like H20 + H20)
Dipole-Dipole Interactions
between molecules that have permanent dipoles(polar molecules). The positive end of on molecule is attracted to the negative end of another.
Example: When 2 H20 molecules interact their (O-H) bonds interact with one another.
London Dispersion Forces (Van Der Waals)
Are the weakest intermolecular forces and arise from temporary dipoles induced in molecules due to the movement of electrons. All molecules, even on-polar ones, experience London dispersion forces, although they are most noticeable in larger, more complex molecules.
Examples: Helium gas (He), Iodine molecules(I2), methane (CH4)
Ion-Dipole
A type of intermolecular force that occurs between an ion( a charged particle) and a polar molecule ( a molecule with a positive and negative end) .
Example: in water, the positive sodium ion (Na+) is attracted to the negative side of water molecules, while the negative chloride ion (Cl-) is attracted the positive hydrogen side of the water molecules. This helps salt dissolve in water
Hydrogen Bonding
specific type of dipole-dipole interaction that occurs when a hydrogen atom, covalently bond to a highly electronegative atom ( like flourine, oxygen, and nitrogen or FON).
Example: HF, NH3, H2O
Unit 2: Photosynthesis
6 CO2 + 6H2O + Light energy --> C6H12O6 + 6 O2
Chloroplasts: The site of photosynthesis in cells
Photosynthesis
Stage 2: Calvin Cycle
Carbon Fixation
Reduction
Regeneration
G3P is the key molecule product for the Calvin cycle, helping the plant to continue producing glucose.
Stage 1: Light Reactions
Solar energy gets converted to chemical energy.
H2O is split, which provides electrons and H+
O2 is released
NADP+ is reduced to NADPH
Phosphorylation: ATP is generated with addition of phosphate group to ADP
Photosystem II:
-chlorophyll a absorbs at 680 nm
Photosystem I:
-chlorophyll a absorbs at 700nm
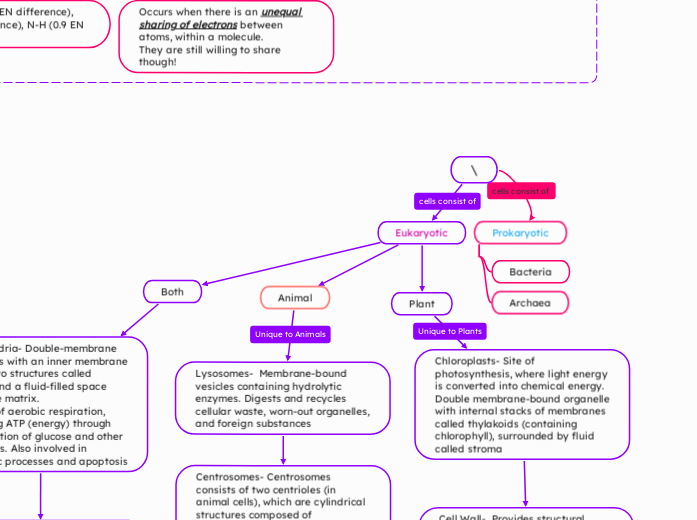
Unit 1: Cells
Prokaryotic
Archaea
No peptidoglycan
Bacteria
Contains peptidoglycan
Eukaryotic
Plant
Chloroplasts- Site of photosynthesis, where light energy is converted into chemical energy. Double membrane-bound organelle with internal stacks of membranes called thylakoids (containing chlorophyll), surrounded by fluid called stroma
Cell Wall- Provides structural support, protection, and regulates cell growth. Rigid outer layer made of cellulose
Large Central Vacuole- Stores water, nutrients, and waste products; provides turgor pressure for maintaining cell structure. Large membrane-bound sac (larger in plant cells)
Animal
Lysosomes- Membrane-bound vesicles containing hydrolytic enzymes. Digests and recycles cellular waste, worn-out organelles, and foreign substances
Centrosomes- Centrosomes consists of two centrioles (in animal cells), which are cylindrical structures composed of microtubules. Involved in organizing microtubules during cell division (mitosis) and forming the mitotic spindle.
Both
Mitochondria- Double-membrane organelles with an inner membrane folded into structures called cristae, and a fluid-filled space called the matrix.
The site of aerobic respiration, producing ATP (energy) through the oxidation of glucose and other substrates. Also involved in metabolic processes and apoptosis
Nucleus- A membrane-bound organelle containing chromatin (DNA and proteins) and a nucleolus, surrounded by a double membrane (nuclear envelope) with pores.
Function: Acts as the control center of the cell, housing genetic material (DNA) and regulating gene expression, growth, metabolism, and cell division