Floating topic
ca
CONCEPT MAP III
DNA REPLICATION
SSB
at
helicase
unzips DNA
Ligase
Glues/connects everything together
DNA Polymerase I
Primase
Synthesizes short RNA primers
Topoisomerase
keeps it from breaking
DNA Polymerase III
Adds nucleotides to the 3' end of a growing DNA strand.
DNA replication reads 3' to 5', makes DNA 5'to3'
Okazaki fragments
Short segments of DNA synthesized on lagging strand
EUKARYOTIC VS PROKARYOTIC GENE REGULATION
Regulation
Occurs at MULTIPLE levels
Control of Gene Regulation also occurs on Epigenetic, post-transcriptional, translational and post-translational level.
Primarily on transcriptional level
Complex, multicellular
Gene Regulation
Key mechanism: Operons
Repressible Operons
TRP Operon: Off when tryptophan is abundant
Inducible Operons
EXAMPLE: LAC Operon, ON when lactose is present
s
Simple, single celled
Translation
Elogation
mRNA enters
added from N to C
E site
Empty tRNA from P site leaves
P site
A site
forms peptide bonds between amino acids attached to tRNA
read from 5' to 3'
Stop codon enters A site
Release factor comes to A site instead of tRNA
Initiation
fMet is starting amino acid
Met is starting amino acid
mRNA, tRNA, Ribosome
Transcription
RNA polymerase II
Transcription factors
RNAP
Unwinds DNA
Creates new pre-RNA transcript
RNA splicing
Splicesome removes introns
Creates new RNA transcript
Termination
Reads until AAUAAA sequence and adds A polytail at the 3' end
Terminates at given termination site
reads 3' to 5'
Promoter
Mutations
Lead to genetic variation
Random Changes in DNA structure
Point Mutations
Frameshift
One or more base pairs swapped that alters the reading of the DNA sequence
Substitution
Nonsense mutation
Introduces an early stop codon
Truncation/faulty protein
Missense Mutation
Change in an amino acid in the protein
Potential changes in protein structure and function
Silent Mutation
No change in amino acid coded for
No effect
Redundancy of genetic code, multiple codons can code for the same amino acid
CONCEPT MAP 2
Membrane Proteins (Action Potential)
Integral membrane proteins
Hyrdophobic regions allow them to interact with internal layer
Na+/K+ pump
Resting Potential
Depolarization
"Peak Phase"
Repolarization
Hyperpolarization
Refractory Period
Return to resting potential
Na+, K+, Ca, H+ will travel through these channels
Photosynthesis
Chloroplasts
PSII and PSI
PSI is second to ACT
Production of NADPH
Used in Calvin Cycle
PSII is First to ACT
Generate high energy E for ETC
Light Dependent Reactions
Calvin Cycle
Conversion of CO2 into Glucose
A regeneration phase
G3P is used to regenerate RuBisCO
Is used for carbon fixation
Carbon fixation
G3P formation
Photolysis
Production of O2 as a byproduct
Light absorption by PSII
Transport of H+ through ETC
ATP and NADPH Production
3 ATP:1 molecule NADPH
Cellular Respiration
Pyruvate Oxidation and Citric Cycle
Pyruvate Oxidation
Pyruvate transported to mitochondria
Pyruvate (3-carbon) is decarboxylated, meaning it loses a carbon atom as CO₂
Coenzyme A (CoA) binds to the remaining 2-carbon fragment, forming acetyl-CoA.
NAD⁺ accepts electrons and a proton (H⁺), becoming NADH
1 Acetyl-CoA, 1 NADH, 1 CO₂
Glycolysis
Oxidative Phospholyration
Products: 3 NADH, 1 FADH₂, 1 GTP (or ATP), 2 CO₂
ETC
NADH donates electrons to Complex I, FADH₂ donates electrons to Complex II.
Protons (H⁺) are actively pumped into the intermembrane space by Complex I, Complex III, and Complex IV creating gradient
The protons flow back into the mitochondrial matrix
The flow of protons through ATP synthase drives the conversion of ADP and inorganic phosphate (Pi) to ATP
Mobile electron carriers: Ubiquinone (CoQ) and cytochrome c
Protein complexes I, II, III, and IV.
ATP, Water, NAD⁺ and FAD (recycled electron carriers that go back to glycolysis, pyruvate oxidation, and the citric acid cycle).
Products: 2 ATP, 2 NADH, 2 Pyruvate, Water
Glucose (C₆) is converted to glucose-6-phosphate (G6P) by hexokinase
Glucose-6-phosphate (G6P) is converted to fructose-6-phosphate (F6P)
Fructose-6-phosphate is converted to fructose-1,6-bisphosphate (F1,6BP) by phosphofructokinase
Cell Signaling
G Protein Receptor
cAMP
AMP
phosphatase
GDP
GTP
3 Stages
response
Transduction
Reception
Intracellular
membrane receptors
Ion Channel Receptor
voltage
ligand
stretch
Tyrosine Kinase
G-protein linked
long distance
hormone
local signaling
synaptic
paracine
Membranes
Function
Cell Recognition
Ligands attaching to membrane receptors
Selectively Permeable
Regulates what enters/exits cells
Components of Membranes
Carbs
Attached to lipids or proteins (glycolipids/glycoproteins)
Cholesterol
Membrane Proteins
Peripheral Proteins
Loosely attached to surface of membrane, assist with signaling and structure
Integral Proteins
Transmembrane, meaning they span the length of the membrane, involved in transport, signaling
Phospholipid Bilayer
Hydrophilic Head
Hydrophobic Tail
Chemical Bonds/Interactions
Intramolecular
Ionic Bonds
When one atom donates one or more of its electrons to another and gain a positive charge due to the loss of an electron. The receiving atom gains a negative charge.
Hydroxide group (OH-)
Sodium Chloride (NaCl)
Carboxylate group (COO-)
Sulfate group (SO4^2-)
Covalent Bonds
Non-polar
Some molecules that have a difference greater than 0.5 can be non-polar due to linearity
Carbon dioxide (CO2)
When there is an electronegativity difference less than 0.5 between the atoms of a molecules
Methyl group (CH3)
Oxygen (O2)
Polar
When there is an electronegativity difference greater than 0.5 between the atoms of a molecules
Sulfhydryl group (SH)
Hydroxyl group (OH)
Water (H20)
Intermolecular
Dipole-Dipole Interaction
Hydrogen Bonds
Special type of Dipole-dipole interaction involving H atoms
Hydrogen can form these bonds with electronegative atoms like oxygen, nitrogen, or fluorine
Positive end of one polar molecule attracted to the negative end of another polar molecule
Ion-Dipole Interaction
When an ionic compound interacts with a polar molecule. Ions are attracted to oppositely charged end of a polar molecule
Salt dissolving in water
Van der Waals/Hydrophobic Interactions
Hydrophobic tails of molecules clump on the "inside" while the hydrophilic heads are "outside" trying to interact with the liquids
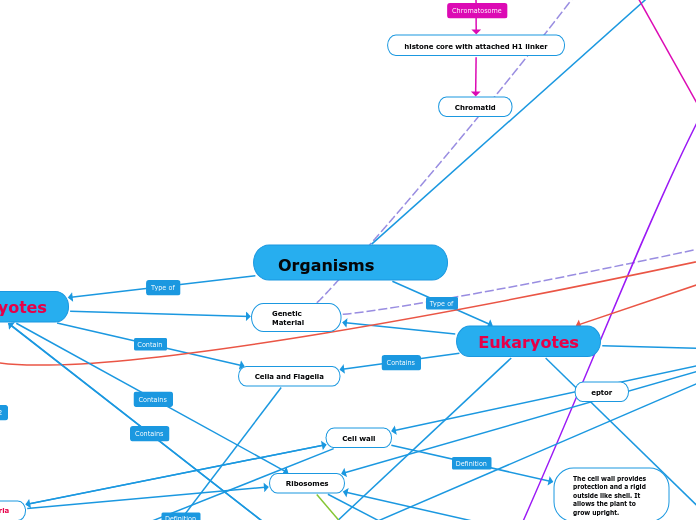
2 Domains of Life (Eukaryotic and Prokaryotic Structures)
Peroxisome
Microfilaments
Microtubes
Mitochondria
Ribosomes
Nuclear Envelope
Nucleus
Plasma Membrane
Smooth ER
Rough ER
Golgi apparatus
Eukaryotes
Intermediate
Gap
Tight Junctions
ECM
Flagellum
Lysosome
Prokaryotes
Central Vacuole
Plasmodesmata
Chloroplast
Cell wall
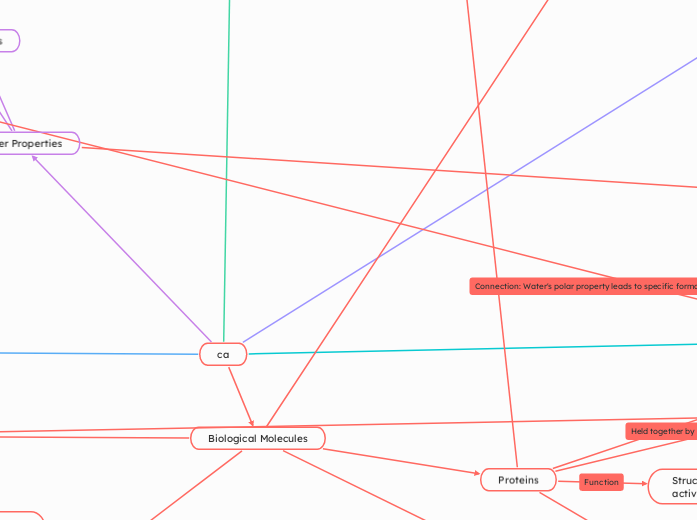
Water Properties
Biological Roles
Higher density as liquid than solid
Allows Ice to float on liquid water
Allows for aquatic environments to be insulated
Universal Solvent
Chemical Properties
Hydrogen bonding
Polar Molecule
Physical Properties
High Specific Heat and Heat capacity
High boiling and melting point
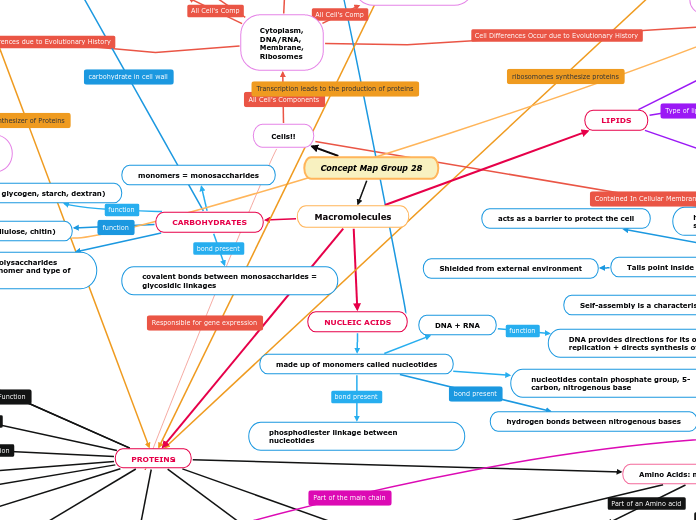
Biological Molecules
Nucleic Acids
Store and transmit genetic information
Nucleotides
Held together by Phosphodiester bonds
Phosphate group, 5 carbon sugar, Nitrogenous base
Proteins
Structural Support, enzymatic activity
Peptide Bonds
Monomers: Amino Acids
Lipids
Functions: Energy Storage (Triglycerides)
Bonds: Ester Bonds
Monomers: Fatty acids and glycerol
Carbohydrates
Functions: Provide energy, structural (cellulose in plants)
Bonds: Glycosidic Bonds
Monomers: Monosaccharides
Examples: Glucose, Fructose, Galactose