arabera Darian Hernandez 2 years ago
430
Biological Molecules
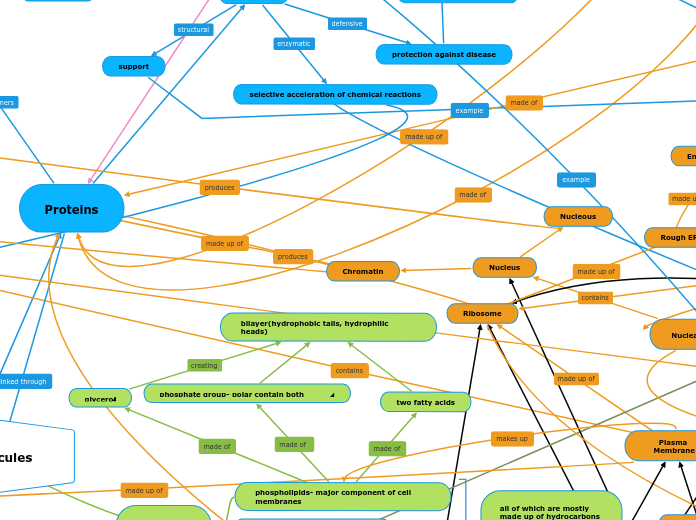
arabera Darian Hernandez 2 years ago
430

Honelako gehiago
Operon ON
Lactose present, Repressor inactive
When there is no Glucose and Lactose is present, then the Operon is ON, because an Inducible Operson (as expression of Lac Z, Lac Y, Lac A genes) is induced by the presence of Lactose
Lactose present, glucose (cAMP level is high), causing an abundant lac mRNA to be synthesized
When lactose is present, the Lac repressor protein binds Lactose, such that it can no longer bind to the operator sequence. RNAP can now bind promoter for transcription of lac operon genes.
The activator protein is present (because an activator protein is required for function) and it's called CAP (Catabolite activator protein). It is activated by cAMP. Adenylyl cyclase forms cAMP from ATP. cAMP binds CAP and CAP helps RNAP to bind promoter to facilitate transcription, so the operon is ON
Operon OFF
Lactose present, Glucose present (cAMP level is low): little lac mRNA is synthesized
If there is no Lactose, then Lac Repressor will be bound to the Operator preventing expression of Lac Operon genes.
The presence of Glucose blocks Adenylyl Cyclase preventing the production of CAMP (if there is no cAMP present then the CAP is inactive, so it can't help the RNAP bind to the promoter.
Lactose absent, Repressor Active
When there is no Lactose, the Operon is OFF, because you need an enzyme. Due to this Transcription, Lac Z, Lac Y, and Lac A is blocked (thus causing the Operon to be OFF).
It's found in E. Coli and is founded by Francois Job and Jacques Monod in 1962. It is an example of both Positive and Negative regulation.
Regulatory Region
Operator
Promoter
Regulatory Gene
Lac I
Stuctural Genes
Lac A
Lac Z
Lac Y
Negative Regulation
When the Repressor protein is bound to the Operator sequence, then the gene expression if OFF
Positive Regulation
The Operon gene expression if ON (expression is at a high level when the activator is bound to the operator)
The activator bind to certain mediator proteins and general transcription factors, helpingthem form an active transcription initation complex on the promoter
Groupings of distral control elements which are far from the gene they control
Repressors
Reduces levels of transcription
Activators
Increases levels of transcription
Additional Transcription Factors bind to the DNA along with RNA Polymerase II, forming the transcription initation complex. RNA Polymerase II then unwinds the DNA double helix, and RNA synthesis begins at the start point on the template strand
The 30-nm fiber that forms looped domains that attach to proteins
Interactions between Nucleosomes cause the thin tiber to coil or fold into this thicker fiber
Linker DNA
The DNA connecting each nucleosome
Histone
Small protein that DNA wraps around
H1
It's not part of the Nuclesome, but is involved in forming the next level of packaging
Histone Core
H4
H3
H2B
H2A
Compacted Chromatid that are made of DNA (genes) and proteins
Chromatid
The fibrous double stranded DNA to which proteins are attached to, once it's compcted it formed Chromosomes
10nm Fiber
DNA winds around histones to form Nucleosomes "beads." They are then strung together like beads on a string by linker DNA
To the left of the transcription start site nucleotides are numbered by negative numbers
Nucleotides in DNA to the right are labeled by positive numbers
sequence AAUAAA is a signal for ribonuclease to make a cut in the newly formed pre mRNA and release it from DNA. At the 5’ end a modified G nucleotide CAP is added, and at the 3’ end a polyA tail is added by polyA polymerase. The 5’cap will be used for translation and the 3’polyA tail helps with stability of the mRNA.
Pre mRNA contains introns and exons. Introns are sequences that need to be removed before translation.
Before the pre mRNA exits the nucleus to be translated it has to remove introns and join together exons. Different genes contain different number of introns.
There is a complex of RNA and proteins called Spliceosome that binds the junctions of the introns and makes cuts to release the introns from the DNA. The exons are then joined together.
Presence of introns help alternate splicing. Different combinations of exons can be generated through removal of different introns to form different mRNAs and hence different proteins.
RNA polymerase II moves downstream, unwinding the DNA and elongating the RNA transcript 5' to 3'. In the wake of transcription, the DNA strands re-form a double helix.
A eukaryotic promoter commonly includes a TATA box (a nucleotide sequence containing TATA) about 25 nucleotides upstream from the transcriptional start point.
Several transcription factors, one recognizing the TATA box, must bind to the DNA before RNA polymerase II can bind in the correct position and orientation.
Additional transcription factors bind to the DNA along with RNA polymerase II, forming the transcription initiation complex. RNA polymerase II then unwinds the DNA double helix, and RNA synthesis begins at the start point on the template strand.
termination
Eventually, the RNA transcript is released, and the polymerase detaches from the DNA.
Translation and protein transport
Termination
Protein transport- all of protein synthesis begins on free ribosomes. The different sequence of amino acids tell proteins what their final location will be and their function.
During translation an SPR will bind to the peptide stand and will momentarily pause the synthesis. The SPR will then also bond itself to a receptor protein found in the membrane of ER.
This SRP will then leave the signal peptide which causes the polypeptide synthesis to resume its natural course. At the same time we have translocation of the protein in the ER membrane.
The protein is then cut by an enzyme found in the receptor. Once this happens the polypeptide is folded to its final conformation.
The protein the goes through further folding in the Golgi, it is shipped here from the ER through a vesicle.
After this the protein is then released into the cell so it cal reach its designated location. Whether it be another organelle like the nucleus or mitochondria, or secreted from the cell.
Once the stop codon is reached a release factor will stand in the A site and this will dissociate the complex stopping translation
A stop codon is either UAG, UAA, or UGA. the dissociating of the complex stopping translation is a GTP driven processes as well.
Elongation
After the initiation process comes the elongation step. In the A site of the large ribosomal subunit a new incoming tRNA base-pairs with the mRNA. Many tRNAs will attempt to bind to the codon in the A site but only the appropriate anticodon will bind to the mRNA.
A peptide bond will then form between the amino acid in the P site and the A site. This bond occurs between the amino acid found in the A site and the carbonyl end of the polypeptide in the P site. This bond removes the polypeptide from the tRNA in the P site and adds it to the amino acid in the A site.
The last step in elongation is called Translocation. In this final step the tRNA shift down a site, meaning that the tRNA in the P is shifted to the E site and is released. The tRNA is shifted to the A site is moved the the E site and at the same time the mRNA iOS moved with its corresponding tRNAs. Meaning that the A site will be open for the next appropriate tRNA to bond.
The translocation step will repeat and repeat until it reaches the stop codon, which then leads to the termination process of translation.
Initiation
At initiation a small ribosomal subunit binds to am mRNA. The small ribosomal unit then scans the mRNA until it finds AUG, which is the start codon.
An initiator tRNA, that consists of an anticodon of UAC will base-pair with AUG. This initiator tRNA carries the amino acid MET, which is methionine.
A large subunit then joins and becomes a translation initiation complex. Translation factors are also used here to bring all the translation components together.
The initiator tRNA is located in the P site of the large ribosomal subunit, while the A site is open to receive the next tRNA with the corresponding amino acid.
An initiator tRNA is made up of a single strand of RNA with about 80 nucleotides. When in three dimension the tRNA is shaped in an upside down L and has the anti codon towards the bottom. On the top side of the upside down L it has a corresponding amino acid
The purpose of the tRNA is to bring the correct amino acid to the mRNA during translation.
elongation
RNA polymerase moves downstream, unwinding the DNA and elongating the RNA transcript 5' to 3'. In the wake of transcription, the DNA strands re-form a double helix.
initiation
After RNA polymerase binds to the promoter, the DNA strands unwind, and the polymerase initiates RNA synthesis at the start point on the template strand.
Primase makes RNA primers complementary to the DNA parent strand. This causes DNA polymerase lll to add nucleotides only to the 3' end
A protein called sliding clamp works with DNA polymerase lll and helps keep it on the parent strand so it does not fall off during replication
along the leading strand the DNA polymerase continuously moves forward towards the replication fork
to form the lagging strand multiple RNA primers are laid down and extended by DNA polymerase lll and making okazaki fragment. the primers are then removed by DNA polymerase l and replaced by DNA nucleotides. an enzyme called ligase seals any gaps by connecting nucleotides with phosphodiester linkages
to connect these nucleotides that are in DNA together we have to use phosphodiester bonds using dehydration/ condensation reactions.
Stomata close during the day, and C O2 is released from organic acids and used in the Calvin cycle
CO2 released into a neighboring sheath cell wherein it is fixed using Rubisco and goes through the Calvin cycle to make sugars
forms a 6 carbon unstable intermediate which splits to form 6 molecules of 3 phosphoglycerate
6 molecules of ATP and 6 molecules of NADPH used to form 6 molecules of G3P
5 molecules of G3P continue on to make more RuBP and 1 molecule of G3P leaves the cycle to form glucose and other sugars
excess NADPH present
only PSI used--as e- are transferred to Fd, instead of forming NADPH they are recruited to the cytochrome complex and plastocyanin molecules of the ETC
movement of electrons leads to formation of ATP by photophosphorylation
Photosystem II
electron transport chain
e- from primary acceptor go down plastoquinone (Pq), cytochrome complex, Plastocyanin (Pc), ferredoxin (Fd)
chlorophyll molecules of photosystem I
formation of ATP by phosphorylation--energy from ETC used to pump H+ into thylakoid space against concentration gradient, and go back down concentration gradient through ATP synthase
photon of light is absorbed by chlorophyll fouind in light harvesting complexes
causes e- to jump to excited state, and go back down to ground state, releasing energy
released energy absorbed by another molecule, and process repeats
main reaction center pair of chlorophyll a molecules (P680) where e- are grabbed by an electron acceptor molecule
splitting H2O (O2 is released)
Photosystem I
photon of light absorbed by chlorophyll causes e- to be excited, as they go back to the ground state energy is released
main chlorophyll a molecules (P700) where e- are grabbed by a primary electron acceptor
electrons go to Ferridoxin (Fd) then on to NADP+ to form NADPH
chlorophyll--light harvesting pigments
When pigments absorb light, an electron is elevated from a ground state to an unstable, excited state
Electrons fall back down to the ground state, releasing photons that cause an afterglow, giving off light and heat
CHO
CH3
Porphyrin ring--light absorbing “head” of molecule; magnesium atom at center
long hydrocarbon tail inserted in the thylakoid membrane
DHAP converts to G3P so at the end we have 2 molecules of G3P and 1 molecule of glucose
G3P id oxidized by transfer of electrons forming NADH a phosphate group is attatched to the oxidized substrate
phosphate group is transferred to ADP and G3P is oxidized to the carboxyl group of an organic acid
enzyme relocates the remaining phosphate group
enolase causes double bond to form in substrate by extracting a water molecule
the phosphate group is transferred from PEP to ADP forming pyruvate
an electron transport chain is used that was energy is released throughout each step
electrons can be directly transferred to Oxygen but this will cause an explosion due to the release of heat and light energy
pyruvate formed in glycolysis enters the mitochondria and is oxidized. The product of oxidation enters the citric acid cycle generating more electron carriers, NADH, FADH2
NADH and FADH2 carries electrons down the electron transport chain and generates ATP through oxidative phosphorylation
occurs in the mitochondria the location of the ETC is in the inner mitochondrial membrane
there are 4 complexes in the ETC complexes 1, 3, AND 4 are H + pumps so they're job is to pump H+ against they're concentration gradient they get this energy because when electrons are transferred down energy is released
H+ in the intermembrane space go back down there concentration gradient through a membrane transport protein called ATP synthase the energy associated with the H+ gradient is used to add an inorganic phosphate to ADP to form ATP
1 glucose can form 30-32 molecules of ATP
Tubulin Dimer
Keratin
Actin
made of beta glucose and connected by glyosidic linkages
help together by hydrogen bonds and form microfibrils
connected through 1-4 glyosidic linkages and alpha glucose monomers
used in animals
isomers
trans isomers having hydrogen on opposite sides of the double bond
cis isomers having hydrogen atoms on the same side of the double bond
Intracellular receptors
Signals that are polar or can not diffuse through the membrane must use this kind of receptors to enter the cell and fulfill its purpose
Membrane receptors
Ion channel receptor
An ion channels receptor will remain close until a signaling molecule binds to it. Once that molecule binds the channel will open.
Once this channel is open specific ions are free to flow into the cell and are able to change the concentration of the cell affecting its function and activity
Once the signaling molecule removes itself from the ion channel, the ion channel closes and ions are no longer free to flow into the cell
Tyrosine kinase receptor
These receptors are made of two polypeptides, each polypeptide has the ability to function as a kinase. As a phosphate groups are added to tyrosines this is referred to as a tyrosine kinase receptor the activated receptor cannot interact with other proteins to bring about a response from the cell
These receptors are used when the molecule is hydrophilic and cannot cross the membrane due to the charge and polarity.
G protein linked receptor
Used by non polar signals since they can easily diffuse through the membrane. This receptor is found inside the cell.
GPCR will receive a signal and bind/touch a G proteins to remove GDP to activate GTP after activated the G proteins will change its shape and detach from the GPCR and move to its next task
The GPCR will move along the membrane and attach self to an enzyme. Doing so the GPCR will alter the enzyme shape and function the enzyme will then be activated and complete the steps to cellular response.
In this process GTP will use energy to activate the receptor in which able to change back to GDP and will move back to original position. The process can then be repeated from the beginning.
Active site is available for two new substrate
Cooperativity
Binding of one substrate molecule to the active site of one subunit locks all subunits in active conformation.
Equilibrium
No net change occurs
∆G = 0
Endergonic
Energy for Cellular Work
Glutamic Acid conversion to Glutamine
∆G for ATP hydrolysis
Energy required, nonspontaneous
∆G > 0
Exergonic
Energy from catabolism
ATP Cycle
Energy released, spontaneous
∆G < 0
∆G = G(final state) - G(initial state)
S = Entropy
T = Temperature in Kelvin
G = Gibbs
H = Total Energy (Enthalpy)
Every energy transfer or transformation increases the entropy of the universe
Energy cannot be transferred and transformed, but it cannot be created or destroyed
Light Energy
Thermal Energy
Extreme thermophiles
Thrive in very hot environments
Extreme halophiles
Live in highly saline enviroments
Plants, survive by capturing energy from the sun in photosynthesis
polypeptides
amino acid sequence determines protein structure
amino acid will go from buried within protein to the surface
amino acid will go from surface of protein to buried inside
denaturation-protein unfolds back into primary structure (no longer biologically active)
renaturation-reverse conditions and test protein function
only peptide bonds remain
quaternary
2 or more polypeptides come together to form a functional protein
tetramer
hemoglobin
trimer
dimer
intermolecular R group interactions
tertiary
intermolecular R group interactions cause polypeptide to fold
ionic bonds
hydrogen bonds
form intramolecular disulfide bonds through oxidation (only covalent bond between R groups)
hydrophobic/van der waals
secondary
intermolecular hydrogen bond between main chain
beta pleated sheets
alpha helices
primary
intramolecular polar covalent peptide bond through main chain
side chain (R group)
hydrophobic
nonpolar
hydrophilic
charged
acidic
basic
polar
main chain
amino group
positive charge
zwitterion in neutral pH within cell
hydrogen
carboxyl group
negative charge
make a phosphodiester bond
one end has a free phosphate group connected to a 5’ carbon of the sugar, while the other end has an OH group connected to the 3’ end of the end sugar. So we call one end of the nucleic acid 5’ end and the other end is the 3’ end.
pyrimidines (C , T) also U in RNA
purines (A, G)
Translation (information from the mRNA is used to make proteins)
Transcription (information in the DNA is used to make mRNA)
alpha isomer the OH group on the bottom of the structure