GENE EXPRESSION
MUTATIONS
nucleotide excision repair
fixes bulge
frameshift
adding or deleting nucleotides in multiples other than three
nonsense
adds a premature stop codon
silent
changes a nucleotide but the protein remains the same
missense
changing a nucleotide which changes the protein
TRANSCRIPTION
Initiation
Elongation
Termination
pre mRNA
RNA Processing
introns are removed from the gene through spliceosomes
make final mRNA
5' cap and poly-A tail
transcription factors
RNA Pol II
nucleus
coupled transcription and translation
RNA Pol
TRANSLATION
mRNA, tRNA, ribosomes, amino acids,
amino acyl tRNA synthetase, peptidyl
transferase, initiation factors,
elongation factors, release factors
rRNA (ribosomal RNA)
large and small subunit
small subunit binds to 5' cap w/ Met
Met scans to find AUG
large subunit binds to AUG
Met starts in the P site
anticodon bonds to the tRNA
amino acyl tRNA synthetase
actual process of translation, adds
proteins onto the tRNA to form a
protein chain until a stop codon is present
80s in Eukaryotes
70s in Prokaryotes
cytoplasm
REPLICATION
Polymerase Chain Reaction (PCR)
denaturation
anneal
elongation
replicates and amplifies DNA
models of DNA replication
dispersive
semiconservative
enzyme: helicase
forms replication bubble
ORI
leading and lagging strand
replication forks
Okazaki fragments
enzyme: SSB
enzyme: topoisomerase
enzyme: RNA primase
enzyme: DNA Pol III
enzyme: DNA Pol I
Ligase
conservative
311C Concept Map
Gene Regulation
DNA Packaging
DNA wraps around Histones
Nucleosomes
30 nm fiber wraps and connects nucleosomes
H1 is needed for fiber
Chromatin
H1, H21, H2B, H3, H4
H2A, H2B, H3, H4 form histone cores
Eukaryotes
Transcription Factors
General Transcription Factors
Low(Basal) levels of Expression
Proximal Control Elements
(close to promoter)
RNA Polymerase II
Repressor
RNA Pol. II can't bind, resulting in
low expression
Specific Transcription Factors
High Levels of Expression
Eukaryotic
Activators
Binding Sites
Distal Control Elements
Enhancers
Prokaryotes
Operons
Trp Operon
-Tryptophan is Present
-Repressor is active
-Tryptophan is not present
-Repressor is inactive
Lac Operon
Lac I
Lac Z
Breaks down Lactose
Lac Y
Permease
Lac A
transacetylase
Off
-Repressor binded to operator
-Repressor "on"
-RNA polymerase can't bind to promoter
On
-Repressor binded to allolactose
-cAMP levels are low
-CAP is not active and RNA Polymerase
cannot bind well to promoter
-Repressor binded to allolactose
-cAMP levels are high
-CAP is active and RNA Polymerase
binds to promoter
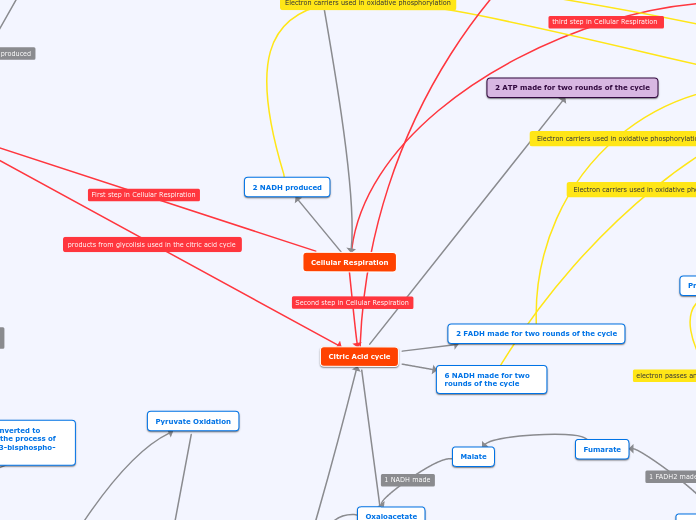
2 NADH produced
Pyruvate Oxidation
pyruvate breaks down and CO2 is released, NADP gets a H+ to make NADPH as the rest of the molecule combines with coenzyme A
Acetyl CoA
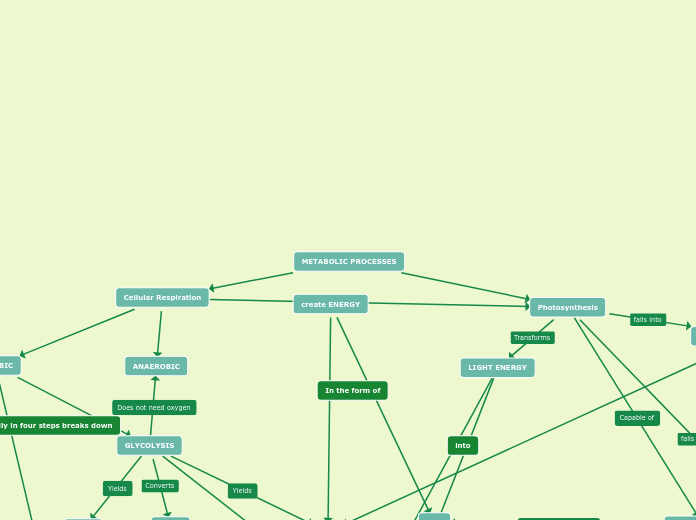
Cellular Respiration
Oxidative Phosphorylation
Chemiosmosis
about 26 or 28 ATP produced
ATP Synthase
The power generated from the flow of protons enables ATP Synthase to add an inorganic phosphate to ADP to form ATP
Electron Transport Chain
The process by which the electrons from the electron carries powers the proton pumps, an H+ gradient is formed with a higher concentration of H+ in the inner membrane (outside). This will provide the potential energy needed in chemiosmosis.
Protein complex 1
NAD+ by product
compex Q
Protein complex 3
Cyt-c complex
Protein complex 4
H2O by product
Protein complex 2
FAD by product
Citric Acid cycle
2 ATP made for two rounds of the cycle
2 FADH made for two rounds of the cycle
6 NADH made for two rounds of the cycle
Acetyl CoA --> citrate --> Isocitrate
a-ketoglutarate
Succinyl CoA
Succinate
Fumarate
Malate
Oxaloacetate
Glycolisis
2 NADPH
2 ATP
Pay-off Phase
G3P converted to 1,3-bisphospho-glycerate
2NAD+ converted to 2NADH in the process of making 1,3-bisphospho-glycerate
1,3-bisphospho-glycerate converted to 3-Phospho-glycerate
3-Phospho-glycerate converted to 2-phospho-glycerate
2-phospho-glycerate converted to phosphoenol-pyruvate
phosphoenol-pyruvate converted to Pyruvate
Investment Phase
Glucose is converted to glucose 6-phosphate
glucose 6-phosphate converted to Fructose 6-phosphate
Fructose 6-phosphate converted to Fructose 1,6-bisphosphate