Photosynthesis
6CO2 + 6 H2O + Energy --> C6H12O6 + 6O2
- H2O is oxidized
- CO2 is reduced
GOAL= Make sugar for energy
Calcin Cycle
- Occurs in Stroma
- Whole Cycle = 9 ATP, 6 NADPH, produces 1 G3P
- 1 Glucose Molecule = 3 CO2, 18 ATP, 12 NADPH
- Makes ATP
Reduction
- 6 NADPH --> 6 NADP+, 6Pi = 6 G3P --> exports 1 (PRODUCT)
Regeneration
- 5 G3P from Reduction, 3 ATP --> 3 ADP = 3 RuBP for Phase 1
Carbon Fixation
- CO2 + RUBP (use Rubisco - ENZYME) = 6C (unstable) = 3C
- 6 ATP --> 6 ADP = 6 1-3 Bisphosphoglycerate
Parts
Chlorophyll
Stromata
Chloroplast
- Site of Photosynthesis
- Found in Mesophyll (interior tissue of leaves)
Photorespiration Adaptations
- ENZYME = Pep Carboxylase
- Photorespiration interferes with Carbon Fixation = adaptations
- Rubisco prefers O2 > CO2 = releases CO2 = blocks photosynthesis = NO ATP/Sugar
C4 Plants
DIFFERENT CELLS
- Mesophyll = CO2 Fixation
- Bundle Sheath Cell = Calvin Cycle
CAM Plants
DIFFERENT TIME OF DAY
- Night: CO2 Fixation, Stromata Open
- Day: Calvin Cycle
Light Reactions
- Occurs in Thylakoid of Cytoplasm
- Makes NADPH Electron Carrier
Cyclic Electron Flow
- PRODUCTS = ATP
- Occurs ONLY when NADPH is in excess
- ONLY uses PS I (P700)
- ELECTRON TRANSPORT CHAIN
Linear Electron Flow
- PRODUCTS = ATP, O2, H+, NADPH Electron Carrier
- ATP goes against gradient in Thylakoid (High H+, Low pH)
- Down gradient through ATP Synthase into Stroma (Low H+, High pH)
- = Photophosphorylation
- Uses PS II (P6800) and PS I (P700)
Glycoprotein
Tags bind to cytosol receptors
Protein ready for secretion
Proteins transport back to Rough ER
Protein transports back to Golgi Apparatus
Protein transports back to Lysosome
Ribosome (rRNA)
Composed of proteins and RNA. Ribosomes (prokaryotic 70S and eukaryotic 80S) have two subunits: large (50S prokaryotic and 60S eukaryotic) and small (30S prokaryotic and 40S eukaryotic) that's about 52 proteins, if prokaryotic, or about 78 proteins, if eukaryotic, attached to mRNA for translation.
Small Subunit
Large Subunit
E site
tRNA exits ribosome
Release factor
Release factor dissociates the ribosome and mRNA is released
Free polypeptide
Polypeptide chain forms to form the protein signaled
Newly formed protein is transferred to the rough ER to refine protein for pathway
Rough ER
Protein Sorting
Plasma Membrane
Extracellular Matrix (Eukaryotes Only)
Secretion
Back to ER
Lysosomes
Membrane Protein
Types of Secreted Proteins
Serum Proteins (Albumin)
Digestive Enzymes (Amylase)
Milk Proteins (Casein)
Extracellular Matrix Proteins (Collagen)
Peptide Hormones (Insulin)
P site
A site
Peptidyl Transferase
Polypeptide chain formed
Concept Map 3
pre-mRNA
poly-A tail
About 50 to 250 adenine nucleotides are added to the 3' end
5' cap
A modified guanine with three phosphate groups is attached to the 5' end of pre-mRNA.
Introns and Exons
Introns are non-coding sequence regions that are removed in RNA splicing.
Exons are genetic coding regions that are combined into one large region for mRNA after RNA splicing.
Alternative RNA Splicing
Sometimes mRNA doesn't need all the exons in the sequence, so introns can be used to extract parts of exons in the sequence to code for different proteins. For example, if there are 7 exons in the chromosomal DNA sequence, but only exons 1, 3, 4, 5, 7 are needed for protein A, then spliceosomes will reduce the mRNA sequence to code for protein A after translation is complete.
RNA Splicing
Spliceosomes can be used to make cuts in pre-mRNA to extract introns to combine exons.
mRNA
Translation
Anti-Codons
tRNA with Amino Acid
Codons
DNA Structure
Single Strand
Nitrogenous Bases
Sugar-Phosphate backbone from 5' to 3'
Double Stranded DNA
Semi-conservative replication
Messleson and Stahl Experiment
The experiment was to use bacterial cells and extract DNA from those cells to predict band distribution. The cells were grown in Nitrogen-15 (high density) combined with CsCl and were spun in a centrifuge for 20 minutes for the first round of replication. The bacteria was then transferred and grown in Nitrogen-14 (normal density) with CsCl and were spun in a centrifuge for 20 minutes for the second round of replication. The experiment concluded that DNA replication was semi-conservative.
Double Helix Structure
Adenine, Cytosine, Guanine, and Thymine
(T and C)
Uracil is also a pyrimidine, but only in RNA.
(A and G)
DNA Mutations
Mutation Sequence &
Consequences
Most Dangerous
Frameshift and Nonsense Mutations are generally the most harmful because they severely disrupt the protein’s structure and function.
Missense Mutations can also be dangerous if they affect a critical region of the protein, such as an active site.
Ironically, I originally wanted to study more into the consequences of Missense mutations and their effects on PrP gene. Prions are an interesting disease that effects proteins and their structure.
This basically causes a protein version of cancer. There is no cure for prion diseases. Commonly this will happen in the brain and the misfolded protein will cause other proteins to fold too
How it Occurs
Spontaneous: Errors during DNA replication not caught by proofreading mechanisms.
Induced: Exposure to mutagens (e.g., UV radiation, chemicals, or carcinogens) damages DNA. Dont smoke cigarettes! This is one of the most common carcinogens!
Types of Mutations
Frameshift
Definition: Insertion or deletion of nucleotides (not in multiples of three), shifting the reading frame of the genetic code.
Consequence: Alters all downstream codons, usually leading to nonfunctional proteins.
Missense
- Definition: Alters a codon, resulting in a different amino acid being incorporated into the protein.
- Consequence: Varies from benign to severe, depending on the location and nature of the amino acid change.
Example in RNA Transcription
CAU -> His
Missense mutation that would change it to
CAG -> Gln
Nonsense
- Definition: Changes a codon to a premature stop codon, leading to failed protein synthesis.
- Consequence: Typically results in nonfunctional proteins or severe loss of function. Also can cause very short proteins
Silent
- Definition: Alters a nucleotide but does not change the amino acid sequence due to the redundancy of the genetic code.
- Consequence: Usually no functional impact on the protein. Hence the word "silent". This happens because the codon wheel has multiple inputs for the same amino acid.
DNA Replication
Process Overview
Process Overview
- Helicase unwinds the DNA, and SSBs stabilize the unwound strands.
- Topoisomerase relieves tension.
- Primase lays down primers.
- DNA polymerase III synthesizes the leading strand continuously and the lagging strand in fragments (Okazaki fragments).
- DNA polymerase I removes primers and fills with DNA
- Ligase seals any remaining nicks to finalize the strand.
Initiation: Helicase unwinds the DNA, primase lays down RNA primers.
Elongation: DNA polymerase synthesizes new strands (leading and lagging).
- Termination: DNA ligase seals fragments; proofreading corrects errors.
Enzymes
Exonuclease
Function: Removes incorrect nucleotides during replication to ensure fidelity.
Interaction: Built into DNA polymerase as a proofreading mechanism.
DNA Ligase
- Function: Joins Okazaki fragments on the lagging strand by sealing nicks in the sugar-phosphate backbone.
- Interaction: Follows DNA polymerase to finalize the lagging strand.
DNA Polymerase III
- Function: Adds nucleotides to the 3' end of the growing DNA strand, using the template strand for complementary base pairing.
- Interaction: Requires RNA primers; synthesizes the leading strand continuously and the lagging strand discontinuously (Okazaki fragments).
Primase
Function: Synthesizes a short RNA primer to provide a starting point for DNA polymerase.
Interaction: Adds RNA primers for polymerase to extend.
Topoisomerase
Function: Relieves supercoiling and torsional strain ahead of the replication fork. Basically Untangles the DNA upstream
Interaction: Works upstream of helicase to ensure smooth unwinding.
SSB Proteins
Function: Stabilize the unwound DNA strands, preventing them from reannealing or being degraded.
Interaction: Coat the strands to keep them separated.
Helicase
Function: Unwinds the double helix by breaking hydrogen bonds between base pairs, creating the replication fork.
Interaction: Opens the DNA so other enzymes can access the strands.
Gene Regulation
- Aims to control expression at earliest stage - usually at Transcription
Regulation
Negative
- Repressor attached = Transcription does not occur
- Repressor unattached = Transcription occurs
Positive
- Activator attached = Transcription occurs
- Activator unattached = Transcription does not occur
Lac I
- Constitutive (Continuous) expression
Repressor Protein
Lac Operon
GOAL = Break down Lactose --> Glucose
- Both positive AND negative regulation
Lactose & Glucose Present
OPERON OFF
- Glucose = Adenyl Cyclase INHIBITOR
- Inhibitor = blocks cAMP product = CAP cannot be activated = RNA Polymerase cannot bind to Promoter
Lactose Present
OPERON ON - POSITIVE REGULATION
- Repressor protein binds LACTOSE
- Needs activator
CAP
- Activated by cAMP (Adenyl Cyclase uses ATP)
- Binds to Promoter to help RNA Polymerase bind
No Lactose
OPERON OFF - NEGATIVE REGULATION
- Repressor protein bind OPERATOR = blocks RNA Polymerase from binding
Operon
Operator
- No lactose = Repressor Protein binds here to turn OFF
Structural Genes
Lac Z
Beta-Galactosidase
- Breaks Glycosidic linkage of Lactose to form Glucose and Galactose
Lac A
Transacetylase
Lac Y
Permease
- Membrane Protein that takes in Lactose
PCE
Proximal Control Element
General Factors
Promoter
- TATA box
- Where RNA Polymerase II binds during bending to control gene expression
DCE
Distal Control Elements/ENHANCERS
Specific Factors
Activators
- Raises transcription above basal levels
- Meets mediator proteins during bending
Repressors
- Lowers transcription to basal/background
- Meets mediator proteins during bending
Transcription
Eukaryotes
RNA Processing
Spliceosome
- ENZYME: cuts introns, joins extrons to form mature mRNA
- KEEPS 5' CAP AND 3' POLY A TAIL
Alternative Splicing
- Removing different introns = different mRNA combinations = different proteins
Transcription
Termination
3' Poly A Tail
- Added by Poly A Polymerase
- Increases mRNA stability
5' Cap
Elongation
- RNA Polymerase II moves downstream of DNA
- Adds to the 3' end using dehydration reactions = double helix
Initiation
TATA Box
- Section on the promoter for Transcription in Eukaryotes
RNA Polymerase II
- ENZYME: binds to the promoter AFTER Transcription Factors bind = Transcription Initiation Complex
- Unwinds DNA and starts RNA synthesis
Transcription Factors
- Binds to the promoter FIRST to help RNA Polymerase II bind next
Prokaryotes
- Occurs in cytoplasm
- Transcription and Translation are coupled = happens immediately
RNA Polymerase
- Enzyme used to make mRNA during Transcription in Prokaryotes
Types of Receptors
Extracellular
Membrane-bound
Intracellular
Concept Map 2
Membrane - Basics
Membrane Fluidity
Membrane Fluidity
- Each phospholipid has a specific phase transition temperature
- Above this temperature, lipids are in a liquid crystalline phase and are fluid
- Below this temperature, lipids are in a gel phase and is more rigid
Membrane Permeability
Low
Permeability
Large, uncharged molecules such as glucose cannot cross on its on
As well as ions like Na+ and K+
Diffusion
Solvent moving from low solute concentration to higher solute concentration. Desire to create an equilibrium.
Facilitated
Diffusion
Facilitated Diffusion is passive transport aided by channel proteins. However, no ATP is required for this type of transport
Plant Tonicity
Animal Tonicity
- Hypotonic: Excess water is in the cell and the cell is lysed and explodes. This is due to high solute concentration within the cell and water rushing inside.
- Isotonic: Water is both leaving and entering the cell in equilibrium. This is also called normal
- Hypertonic: High solute concentration outside the cell causes water to rush outside the cell. Cell gets shriveled up, this usually causes cell death.
High
Permeability
High permeability
- Small nonpolar molecules can cross through on their own
- Small uncharged polar molecules like water and glycerol
Selective
Permeability
Selective permeability of Plasma Membrane
- General rule is the size and charge of the molecule can in and out of the cell
Transport proteins allow the passage of hydrophilic substances across the membrane
Active Transport
- Movement of substances from low to high concentrations (pushing against the concentration gradient)
- Maintains a concentration gradient
- Uses energy (ATP)
Sodium-
Potassium
Pump
The Sodium-Potassium Pump: A specific case of Active Transport
- This pump is present in all animal cells
- This ensures sodium is lower on the inside and more on the outside. This also ensures the potassium is higher on the inside and lower on the outside.
Voltage Difference
Due to these pumps, a voltage difference is made
- Outside the membrane is slightly positive (some more Na+ ions)
- Inside the membrane is slightly negative (some more K+ ions, think of salty banana)
Ion Channels
Voltage-gated
Open and clsoe in response to changes in membrane potential
Ligand-gated
Open and close when a neurotransmitter binds to channel
Stretch-gated
Also known as sense stretched, open when membrane is mechanically deformed
Ungated
Always Open
Carrier
Protein
Pump
Carrier Protein Pump: Carries from protein from inside to outside the cell, uses ATP energy to change shapes within the molecule
Proton Pump
- A protein proton pump uses ATP to pump protons against the concentration gradient
Protons want to go back in so they go down the concentration gradient using facilitated diffusion in a Sucrose cotransporter
- Energy associated with the proton gradient help protons and sucrose move across the membrane
Electrogenic
Pump
- A transport protein generates a voltage across a membrane-membrane potential
- Help stores energy that can be used for cellular work
Plasma Membrane - Outer
Layer of the Cell
Membrane Proteins
Membranes have a variety of proteins
- Peripheral proteins are not fully integrated within the cells
- Integral proteins are integrated fully within the membrane
Some Functions of Membranes Proteins Include:
- Transport
- Enzymatic
- Signal Transduction
- Cell-Cell Recognition
Metabolism/Enzymes
Enzymes
Competitive Inhibior
Competitive Inhibitors bind to the active site of a enzyme prevent substrates from binding to the enzyme and lower overall efficiency of reactions.
Noncompetitive Inhibitor
Binds to another part of the enzyme but affects the shape so that original substrates cannot bind to the new active site
Made of Specialized Proteins
with An Active Site
Enzymes are also pH and Temperature Sensitive.
Most Enzymes have an ideal pH or Temperature that they work at most efficiently, and when these fall outside the parameters the enzyme not might work as well or overall completely denature back to a polypeptide chain.
Overall Goal:
Speed up Chemical Reactions
Lower Activation Energy
of Reactions to Take Place
The Metabolic Pathway
Maintain Homeostasis
Conservation of Energy
Thermodynamics
- System - matter within a defined region of space
- Closed System
- Open System
Laws of Thermodynamics
- First Law of Thermodynamics: Energy can be transferred and transformed, but cannot be created or destroyed.
- Second Law of Thermodynamics: Every energy transfer or transformation increases the entropy of the universe.
Free Energy & Free Energy Change
Free Energy is Cellular Work
Entropy = Gibs Free Energy + T(Enthalpy)
G = H -TS
Measuring change in each at constant of T
Free-Energy Change (ΔG)
- The change in free energy ΔG during a chemical reaction is different between the free energy of the final state and the free energy of the initial state
- ΔG = G(final) - G(inital)
- For a spontaneous process, no energy input is needed - ΔG is negative
- For a nonspontaneous process, energy input is needed ΔG is positive
Ideally in life, we want most reactions to be ΔG<0
Types Include:
Cellular Respiration via
Glucose is oxidized.
Additional:
Anabolic Pathways such as
Polymerization and Photosynthesis
Eukaryotic Cells
DEFINED BY:
- DNA in nucleus
- Double membrane bound organelles
EXTERNAL MEMBRANE:
INTERNAL MEMBRANE:
- Pyruvate Oxidation
- Citric Acid/Krebs Cycle
Animals ONLY
Junctions
Gap Junctions
Desmosomes
- Leaky junctions
- Point of direct contact between cells
Tight Junctions
ECM
- ECM changes = changes in cell
- MNEMONIC: "Can Find Party In ECM rave"
Fibronectin
- Attach ECM to Integrins in Plasma Membrane
Collagen Fibers
- Embedded in web of Proteoglycan complexes
Integrins
- Binds to ECM = Transmit signals between inside and outside of cell
Proteoglycan
- Made of protein + sugar
- Attached to single polysaccharide
Specialized Cells
T Lymphocytes
- T helper cells activate B cells
- B + T interact through surface proteins
- Increased Ribosome, Rough ER, Golgi (for transport)
Macrophage
- Break down + release toxin (engulf + destroy)
- Increased Lysosomes
Lymphocytes
- B Cells = MAKE ANTIBODIES to bind to + destroy antigens
- Increased Ribosomes and Golgi Apparatus (ship out through ER)
Both Plants and Animals
Nucleus
- Site of Transcription + DNA
- DISEASE = Progeria
Nuclear Pores
- Helps transport in and out of nucleus
Nucleolus
Nuclear Lamina (TEM)
Cytoskeleton
- Cell Storage + Organelle Anchorage
Intermediate Filaments
- MORE PERMANENT
- Maintains cell shape, anchors nucleus + organelles, forms nuclear lamina
- Made of KERATIN Proteins
Microfilaments
- THINNEST = made of 2 intertwined actin protein strands
- Actin Filaments
Cytoplasmic Streaming
- LOCALIZED
- Myosin contract in PLANTS = Propels Cytoplasm = Distribution of nutrients
Amoeboid Movement
- LOCALIZED
- Microfilament + Myosin = Tail squeeze interior fluid = Extending Pseudopodium
Muscle Contraction
- Myosin head + Actin filament attach and move = actin filament slides = underlying muscle fibers contract
Microtubules
- ALL EUKARYOTES HAVE
- THICKEST (Alpha and Beta Dimers)
- Contains:
- Dynein = Motor protein to drive bending movement of organelles
- Basal Body = Ring of 9 triplets to anchor
- Cilia + Flagellum
- DIFFER in arrangement of microtubules
Mitochondria
- Site of cellular respiration
- Contains:
- Intermembrane Space
- Cristae: Membrane folds
- Mitochondrion Matrix = Free ribosomes + DNA
- Involved in Cell Respiration to make ATP = POWERHOUSE OF CELL
- DISEASE = Myoclonic Epilepsy
Ribosomes
Free Ribosomes
- Present in CYTOSOL
- Makes Proteins
Bound Ribosomes
- Present OUTSIDE ER
- Protein inserted in membrane/secreted out of cell
Endomembrane System
- Responsible for protein traffic + metabolic functions
Nuclear Envelope
- Regulates transport in + out of nuclear pores
Lysosome
- H+ pump maintains LOW PH INSIDE to keep enzyme functional = Hydrolysis/breaks covalent bonds
- DISEASE = Build up of toxins
Autophagy
RECYCLE
- Hydrolysis enzyme recycles organic materials
- Digests damaged organelles --> Cytosol for reusage
Phagocytosis
EAT
- Cell extends membrane --> Engulf ---> Pinch off
- Digestion products (amino acids + simple sugars) --> Cytosol as nutrients
Vacuoles
Contractile Vacuole
- Pumps excess water out of Freshwater Protists
Central Vacuole
- IN PLANTS ONLY
- Stores Potassium, Water, and Chloride
Food Vacuole
- Formed when Phagocytosis occurs
Golgi Apparatus
- Shipping Vesicle
- Contains:
- Cis Face = Receiving side
- Trans Face = Shipping side
Endoplasmic Reticulum
- Contains:
- Smooth ER (NO Ribosomes) = Makes Lipids, metabolizes Carbs, detoxify
- DISEASE = JAUNDICE
- Rough ER (Bound Ribosomes) = Folds Proteins, secretes Glycoproteins
Plants ONLY
Cell Wall
- Made of cellulose
- Maintains cell shape
- Contains:
- Secondary Cell Wall: Between plasma membrane and primary cell wall
- Primary Cell Wall: Thin + flexible
- Middle Lamella: Between primary walls of adjacent cells
Plasmodesmata
- Cell junction in plants
- Provides channels for water and nutrients to travel between cells
Chloroplast
- Site of plant photosynthesis
- Contains:
- Stroma = Internal fluid
- Thylakoid = Membrane sacs
- Stacks of Thylakoid = Granum
Cell Signaling
Notes
Long-Distance Signaling
Hormonal Signaling
Steroid Hormone Receptors
The steroid hormone, aldosterone, passes through the plasma membrane. Aldosterone binds to an intracellular receptor protein in the cytoplasm, activating it. The hormone-receptor complex enters the nucleus and binds to specific genes. The bound protein acts as a transcription factor, stimulating the transcription of the gene into mRNA. The mRNA is translated into a specific protein.
Epinephrine
One steroid hormone can have multiple effects on cells. If can have the same receptor but different intracellular proteins to activate, or it can have different receptors.
Local Signaling
Paracrine Signaling
Local regulators (or signals or ligands) diffuse through the extracellular fluid to get to the target cell.
Signaling Molecule
Intracellular Receptors
Membrane Receptors
Ion Channel Receptor
Ion channel receptors often have ligand-gated receptors in which the channel will remain closed until a ligand binds to the receptor. When the ligand binds to the receptor and the channel opens, specific ions can flow through the channel and rapidly change the concentration of that particular ion inside the cell. This change may directly affect the activity of the cell in some way. When the ligand dissociates from this receptor, the channel closes and ions no longer enter the cell.
Ligand binds to receptor
Channel opens allowing ions to flow down the concentration gradient
Ions enter the cytoplasm and trigger a cellular response
Ligand dissociates and channel closes
G-Protein-Linked Receptor
Phosphorylation Cascade
Here, we'll look at an example of the amplification effect caused by binding epinephrine to the receptor and causing millions of molecules to be made.
Epinephrine binds to G-Protein-Linked Receptor
Inactive G-Protein is activated and slides across the membrane to bind and activate Adenylyl Cyclase
The ligand can stay on the receptor and continue the amplification effect or dissociate from the receptor and end the effect
After the G-Protein activates Adenylyl Cyclase, it can continue to activate the enzyme or deactivate by delinking a phosphate group from GTP and making GDP and shift back to it's origin
ATP is used to activate Adenylyl Cyclase
Activated Adenylyl Cyclase converts ATP to Cyclic AMP (cAMP) as a secondary messenger
cAMP activates a series of Protein Kinases
Protein Phosphatase (PP) removes a phosphate group to deactivate the proteins
Phosphodiesterase (PDE) deactivates cyclic AMP (cAMP) and converts it to AMP
Cellular Response (millions of molecules)
Synaptic Signaling
Electrical signals along nerve cells trigger a release of neurotransmitters that diffuse across the synapse to reach the target cells so that they can become stimulated and get the signal.
Fermentation
- Occurs in cytoplasm, without Oxygen (Anaerobic)
- RECYCLING PROCESS
- Recycles NADH into NAD+ to generate more Glycolysis
Lactic Acid
- Start: 2 NADH + 2 Pyruvate
- End: Ethanol + Carbon Dioxide
Alcohol
- Start: 2 NADH + 2 Pyruvate
- End: Lactate
Cellular Respiration
- CATABOLIC REACTION (exerting energy)
- Electron Carriers: NADH and FADH2
- Glucose = Oxidized, Oxygen = Reduced
Citric Acid Cycle
- Occurs in the Mitochondria
- 1 Acetyl CoA + 1 NADH = 1 ATP, 3 NADH, 1 FADH2
- Start: Isocitrate
- End: Alpha Ketoglutarate
- Start: Oxaloacetate
- Add: Acetyl CoA
- End: Citrate
Oxidative Phosphorylation
- Occurs in Mitochondria
- Goal is to generate ATP
Chemiosmosis
- H+ traveling AGAINST gradient from ETC supplies energy source to convert ADP to ATP as it travels DOWN to Chemiosmosis
- ATP Synthase is involved
- Synthase grabs protons pumped out as electrons travel down ETC - Facilitated Diffusion
Electron Transport Chain
- Electrons from NADH travel through Complexes I, Q, III, C, and IV down ELECTRON TRANSPORT CHAIN
- Complexes I, III, and IV pump out H+ into the intermembrane space
- Energy from electrons transferring down ETC is used to pump H+ AGAINST concentration gradient
- Once electrons reach Oxygen, water is formed
Pyruvate Oxidation
- Needs Oxygen (Aerobic)
- Pyruvate from Glycolysis gets oxidized in the Mitochondria if O2 is present
- Products go through Krebs/Citric Acid Cycle
- 1 Pyruvate = 1 Acetyl CoA + 1 NADH
Glycolysis
- Starts with electrons from food (glucose)
- Occurs with or without oxygen (anaerobic and aerobic)
- Energy Investment Phase
- Uses 2 ATP
- Creates 2 G3P
- Energy Payoff Phase
- Makes 4 ATP
- Everything is doubled
Output
- "I take a NAP @ 2PM"
- NET: 1 Glucose = 2 NADH, 2 ATP, 2 Pyruvate
Step 3
- Start: Fructose 6P
- Add: Phosphofructokinase
- End: Fructose 1-6 Bisphosphate
Step 1
- Start: Glucose
- Add: Hexokinase
- Removes Phosphate group from ATP
- End: Glucose 6P + ADP
Plant Cells Only
Prokaryotic - The Basics
Archaea - Basics
Archea is rumored to be one of the first-ever cells of life. They are extremely basic, yet this simple structure provides the basis for a stable organism and some unique properties.
- Some Archea live in extreme environments, these are called extremophiles
- Extreme Halophiles - Can survive in high-solute solutions
- Extreme Thermophilies - Can survive in extreme temperatures
- Acidophiles - Can survive in acidic solutions
Archaea Metabolism
Some Archaea are Methanogens, which live in swaps and marshes. These produce Methane a waste product of their metabolism. This provides those environments with a "rotten" smell.
Bacteria - Basics
Bacteria are small simple cells that consist of a fundamental Cell Structure.
These usually consist of
- Cytoplasm
- Membrane
- Chromosomes folded within a Nucleoid
- A small collection of DNA in certain Bacterial cells have a Plasmid.
- Plasmids can also be exchanged between Bacterial cells. This is how antibiotic-resistant genes are transferred between Bacterial Cells.
- Cell Walls
- Uniquely Cell walls are made from Peptidoglycan instead of Cellulose & Beta-Glucose in plant cells.
Certain Bacteria also contain:
- Slime layers that are made of Polysaccharides
- Some Bacteria have Flagellum to propel through solvents like water. Motile function.
- Some have fimbriae and pili that attach to surfaces. hey can also use pili to connect to bacteria and transfer DNA - Plasmid Anti-biotic Resistance
- Some can form endospores which is a hibernation almost where Bacteria can survive in harsh conditions for centuries.
Other minor organells:
- Gas Vacuoles
- Ribosomes
- Inclusion Bodies
- Nucleoids
- Periplasmic Space
Bacterial Metabolism
There are four major nutritional modes of Prokaryotes
Autotrophic Method
- Photoautotrophs
- Chemoautotrophs
Heterotrophic
- Photoheterotrophic
- Chemoheterotrophic
Roles of Oxygen in Metabolism
- Obligate aerobes
- Requires Oxygen for Cellular Respiration
- Obligate anaerobes
- Uses fermentation and anaerobic respiration. Poisoned by Oxygen.
- Facultative anaerobes
- Uses Oxygen when present. Carry out fermentation or anaerobic respiration when Oxygen is not present.
Differences Between Kingdoms
While the Kingdoms might be similar, there are a few major differences, like their organelles:
- Both have the presence of cell walls.
- Both do not have a nucleus
- Archaea has branched lipids that are uniquely made with ether bonds. Bacteria do not.
- Bacteria have the presence of peptidoglycan, whereas Archaea does not.
Andrew
Lipids
Function
Lipids are one of the most important molecules for cells to have function:
- Cellular Membrane: Lipids are fundamental components of cell membranes. Usually phospholipids, these form a structure having hydrophilic facing the outside of the cell and the inside surface of the cell, with the hydrophobic hydrocarbon tails between the membrane.
- Energy Storage: Lipids serve as a dense source of energy for cells. In animals, fats like triglycerides are stored in cells to provide energy when glucose levels are low
- Some other functions that are not covered are insulation/protection for macroscopic organisms, Signaling Molecules, and Electron carriers in cellular respiration.
Structure
Lipids are made from building blocks of sugar:
- One - Monosaccharides
- Two - Disaccharides
- Three - Trisaccharides
- Four or More - Polysaccharides
These saccharides link through the Glycolic Cvalend bond - meaning water performs a hydrolysis reaction that breaks this bond. Fructose and Glucose come in contact and allow water to use hydrogen bonds.
Types of Polysaccharides
- Storage Polysaccharides
- Glycogen
- Starch
- Dextran
- Structure Polysaccharide
- Cellulose
- Chitin
Glycerol fatty acids can have multiple chains formed through ester bonds. Tails that contain only a single bonded chain are called saturated fats. Tails that contain double bonds are called unsaturated fats
Trans geometric structures cases have the hydrogens on both sides of the double bond
Cis geometric structures cases have the hydrogen on the same side of the double bond; this results in a large bend on the hydrocarbon tail.
Eukaryotic - The Basics
To not go too much into structure, this is just the basics of Eukaryotic cells. Eukaryotic cells are very complex cells that have different organelles across different cell types.
Common Organelles:
- Nucleus - A complex envelope that contains Deoxyribose Nucleic Acid that contains the instructions for the cell for its function and the template to create amino acids.
- Mitochondria: Often called the "Powerhouse" of the cell, this is where the synthesis of ATP is done.
- Rough and Smooth Endoplasmic Reticulum - A network of membranes involved in protein synthesis and folding. Rough ER has ribosomes embedded within it. Smooth ER is involved in Lipid synthesis.
- Golgi Apparatus - Modifies, sorts, and packages proteins and lipids for transport.
- Lysosomes: Contain enzymes that break down waste products and cellular debris through the acidic interior of Lysosomes.
- Ribosomes: Small structures that are responsible for protein synthesis through tRNA and rRNA.
- Plasma Membrane: The outer boundary of the cell, and it regulates what enters and exits the cell.
- Chloroplasts: Organelles that capture sunlight for photosynthesis for glucose.
These are pretty common organelles but multiple Eukaryotes have cell-exclusive organelles:
- Flagella for movement through fluid
- Pili for attachment
- Axons, Mylean Sheeth, etc. for nerve cells.
Differences Between Eukaryotic & Prokaryotic Cells & Endosymbiotic Theory
Prokaryotic - Basic small cells with a limited number of organelles
Eukaryotic - Complex larger cells with the presence of membrane-bound organelles
Endosymbiotic Theory
Prokaryotes were the first cells with Eukaryotes coming after. The proposed theory states that a protoeukaryotic cell absorbs a prokaryotic cell to make Mitochondria, and an autotrophic prokaryotic cell makes Chloroplasts.
Evidence for the Endosymbiotic Theory
- The inner membranes of mitochondria and plastid are similar to the plasma membranes of living bacteria
- Replication of mitochondria and plastids is similar to cell division in Bacteria
- Mitochondria and plastids have circular DNA like Bacteria
- Mitochondria and plastids transcribe their own DNA into proteins. There are Ribosomes in mitochondria - almost having an independent cell structure
Joey
Nucleic Acids
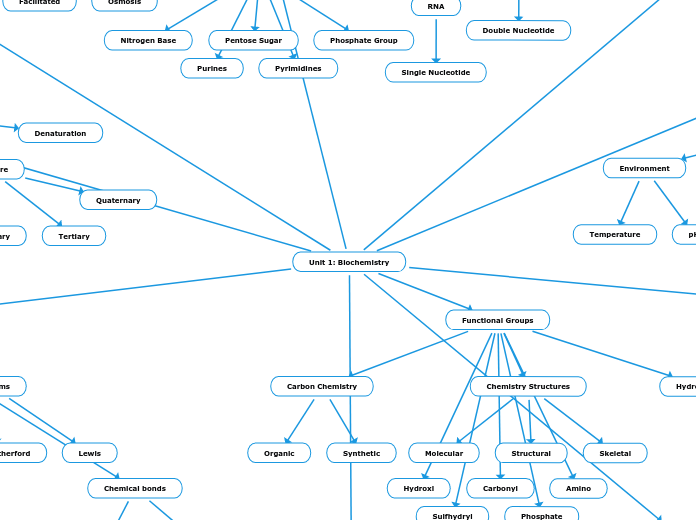
RNA
- Oxygenated pentose sugar with a phosphate group and nitrogenous bases (A, U, C, G)
- Alpha bent structure making it easy for replication and systemization of proteins
- No hydrogen bonds with nitrogenous bases and phosphodiester linkages
DNA
- Deoxygenated pentose sugar with a phosphate group and nitrogenous bases (A, T, C, G)
- Double helix structure easy for stability and replication
- Hydrogen bonds with nitrogenous bases and phosphodiester linkages
Bonds
Intramolecular
Colavent
- Covalent - electrons shared between two atoms under the same electron cloud (ΔEN >1.5)
Polar
- Polar covalent - electrons unequally shared between two atoms (ΔEN 0.5<x<1.5)
Nonpolar
- Nonpolar covalent - electrons equally shared between two atoms (ΔEN <0.5)
Ionic
- Ionic - electrons transferred from one atom to another producing a cation and anion
Intermolecular
BETWEEN MOLECULES
Ion Dipole
- Between ions and polar molecules
- 1 fully charged and 1 partially charged (ie Na+ and Cl- interacting with H2O partial charges)
- Positive charge interact with negative charge
Hydrophobic
- Non Polar parts group together in Polar substance (Water)
- NON POLAR INTERACTION
- Seen in Phospholipid tails
Van der Waals
- Asymmetric electron distribution in molecules
- Electrons pull towards partial positive side
- NON POLAR INTERACTION
- Happens everywhere
Hydrogen
- Hydrogen bonding with F, O, N of adjacent molecule
- POLAR INTERACTION
Carbohydrates
Carbohydrates are organic molecules that serve as energy fuel for short-term storage. Carbohydrates are one of the biomolecules that contain monomers and polymers. The monomers of carbohydrates are called monosaccharides while the polymers of carbohydrates are polysaccharides.
Carbohydrate skeletons can be drawn in four structures: linear, double bond position, branching, and rings.
Monosaccharides
Monosaccharides are monomers that are simple structures like glucose, fructose, and galactose.
Polysaccharides
Polymers are synthesized through dehydration/condensation. Polymers are broken down via hydrolysis and enzyme catalysts.
There are different types of polysaccharides: storage and structure.
Structure Polysaccharides
Storage Polysaccharides
Storage polysaccharides
Glucose
The difference between all the types of polysaccharides is the type of linkages they have and whether or not they are alpha glucose or beta glucose.
Amylopectin
Amylopectin uses alpha 1-6 glycosidic linkage to form branched helix chains. Amylopectin is soluble in water and does produce a gel when hot water is present.
Amylose
Amylose uses alpha 1-4 glycosidic linkage to form helix chains. Amylose is not soluble in water and doesn't produce a gel when hot water is present.
Cellulose
Cellulose is a beta glucose molecule and to make the structure of cellulose found in the cell membrane, it uses beta 1-4 glycosidic linkage. Enzymes cannot be used to break down the linkages, so it is not digestible. Another type of structure polysaccharide is chitin, which is also found in plant cells.
Glycogen
What we would think of when we think glucose. Glycogen is a large, formed polysaccharide that is used by the body as fuel. It uses alpha 1-6 glycosidic linkage to branch out the glucose to store energy to use for fuel.
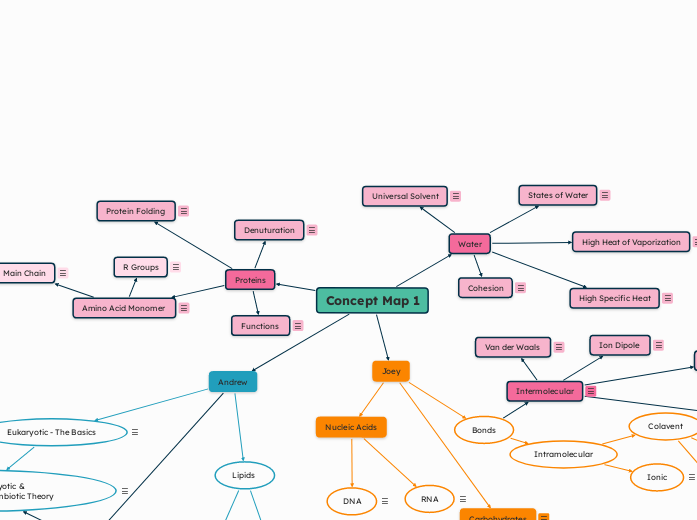
Concept Map 1
Proteins
Functions
- Enzymatic Proteins = Accelerates selective chemical reactions
- Defensive Proteins (ANTIBODIES) = Protection against disease
- Storage Proteins = Stores amino acid monomers
- Transport Proteins = Transports substances through cell
- Hormonal Proteins = Coordinates an organism's activities through secretion of hormones (ie Insulin)
- Receptor Proteins = Helps with cell response to chemical stimuli
- Contractile + Motor Proteins = Cell movement (ie Myosin and Actin in muscles)
- Structural Proteins = Cell support (ie Collagen and Keratin)
Amino Acid Monomer
- Central Carbon bonded to 4 DIFFERENT groups = Enantiomer Isomer (mirror images)
- ONLY L FORM OF ENANTIOMER PRESENT
- Link together through dehydration to form Polypeptides
- Protein functions as Trimer = 3 Polypeptides --> Highest structure = Quaternary
- Protein function as Dimer = 2 Polypeptides --> Possible structures = Primary, Secondary, Tertiary, Quaternary
Main Chain
- Main Chain = Amino (NH2+), Carboxyl (COOH-), and Hydrogen
- SAME FOR ALL AMINO ACIDS
R Groups
- Side Chain = R Groups
- Non Polar (C-H): Hydrophobic and Van der Waals interactions
- N-H can be both non polar or polar but is GENERALLY MORE NON POLAR
- Polar (O-H, S-H, C=O): Polar Covalent interactions
- Acidic: Negatively charged
- Basic: Positively charged
Glycine is NOT an Enantiomer because there are not 4 unique groups around central Carbon
Denuturation
- Environment alters/heats = Protein cannot retain shape = UNFOLDS back to PRIMARY STRUCTURE
- Breaks all bonds EXCEPT PEPTIDE BONDS
- Renature (remove denaturing condition) is successful IF the protein retains original function -- ie cool protein if heat caused denature
Protein Folding
- Primary Structure: Long chain of amino acids
- Involves MAIN chain of aminos
- PEPTIDE Bonds are present
- Secondary Structure: Alpha helicase or Beta pleated sheets
- Involved MAIN chain of aminos
- HYDROGEN Bonds are present
- Tertiary Structure: Polypeptide fold into 3D shape
- Involves R GROUP
- ALL Bonds are present
- Disulfide, Ionic, Hydrogen, Hydrophobic, and Van der Waals
- Quaternary Structure: 2+ Polypeptides form functional protein
- Involves R GROUP
- ALL bonds are present
Water
High Heat of Vaporization
- Amount of heat/kinetic energy absorbed is high enough to break hydrogen bonds and NOT reform
- Evaporation Cooling = stabilizes temperature in water and organism by cooling surface after water evaporates
High Specific Heat
- Helps moderate temperature - defined by amount of heat required to raise temperature by 1 degrees
- Water absorbs heat --> hydrogen bonds break --> water releases heat and bonds reform = water is resistant to changes in temperature
Cohesion
- Allows for water movement against gravity in plants due to hydrogen bonds between water molecules
- Causes high surface tension in water - makes it difficult to break water surface
Universal Solvent
- Dissolve = water seeps inside crystal lattice and surrounds each ion (hydration shell) to break bonds
- Ionic compounds dissolve in water through Ion-Dipole interactions (ie salt)
- Polar compounds dissolve inw ater through Hydrogen bonds (ie sugar)
- Water SURROUNDS Nonpolar compounds because they are Hydrophobic
States of Water
- Solid state = hydrogen bonds form and DO NOT break
- Strong, ordered crystal lattice structure
- Expansive upon freezing
- Liquid state = hydrogen bonds break and reform constantly
- (Denser state than solid
- Gas state = hydrogen bonds break and DO NOT reform