da Jacob Findlay mancano 3 anni
207
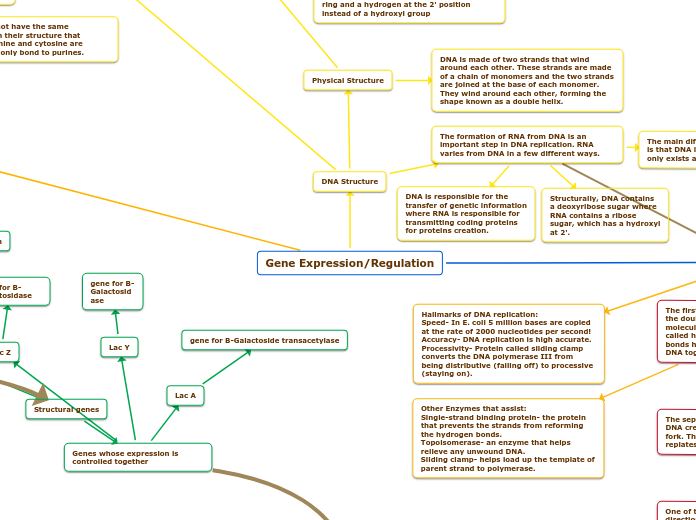
Gene Expression/Regulation
The organization of genes involves structural genes and regulatory genes, with structural genes like Lac Z, Lac A, and Lac Y being responsible for various functions in gene expression.
da Jacob Findlay mancano 3 anni
207

Più simili a questo
gene for B-Galactoside transacetylase
gene for B-Galactosidase
Binding causes positive/negative regulation
activators/repressors
activator bind to mediator proteins
brings the activator closer to the promotion
DNA bending brings activator closer to promotion site
activator binds to enhancer
turns gene regulation on/off
last gene that enters the nucleus
repressor
operon gene regulation on
activator
Transcription factors
help increase or decrease level of transcription
General
background/basal
low levels of transcription
bind to promoter and regions near
Specific
changes levels of transcription
increase levels of transcription
done by activators
high levels of transcription are reduced by repressors
Bind to distal control elements called enhancers
Present near or far from gene they are controlling
control elements in DNA
proximal
sequences in DNA close to promoter
Bind general transcription factors
distal
sequences in DNA upstream or downstream
can be close or far from gene they are controlling
Bind to specific transcription factors
operons
helps with regulation with an on-off switch
"switch"
segment of DNA known as "operator"
positioned within promoter
proteins bind to operators to turn on gene expression for multiple genes
OR to turn off expression
positive regulation
occurs at level of transcription
gene expression on
with activator, transcription occurs
expression at high level
gene expression off
transcription occurs
no transcription occurs
repressor protein bond to operator sequence
Lac operon
regulation needs both repressor and operator
lactose
disaccharide made of glucose and galactose
inducer of the lac operon
cAMP level high
abundant lac mRNA synthesized
operon on: induced/high expression
Activator protein CAP is activated by cAMP
CAP helps RNAP bind to promoter
facilitates transcription
negative regulation
transcription of structural genes is blocked
operon on
mRNA translates: B-Galactosidase, Permease, transacetylase
takes in more lactose from outside
breaks it down to glucose and galactose
uses sugars as needed
all structural genes are transcribed
forms a long mRNA
no glucose = operon on
inducible operon
cAMP level low
operon off
blocks adenylyl cyclase
prevent production of cAMP
CAP cannot be activated
CAP can't help RNAP bind promoter
little mRNA synthesized
transcription factors (instead of operons)
activators
repressors
Operons do not occur in eukaryotic cells - made in mRNA w/ individual promoter
Distal
enhancers
bind activator proteins
activator bound to receptor is brought to promoter via DNA bending proteins
Transcription increased via RNA polymerase II
Proximal
basal (general) expression
Specific to eukaryotes (ex. humans)
The monomers of DNA are nucleotides. Nucleotides are made up of three parts: a sugar, a base, and a phosphate.
A phosphate group is a functional group made of one phosphorus atom bonded to four oxygen atoms. Each DNA monomer has one phosphate group attached to it.
A base is a nucleotide found in the "rungs" of the DNA double helix ladder. These bases are connected by hydrogen bonds.
DNA contains a deoxyribose sugar. A deoxyribose sugar is formed by a 5-carbon ring and a hydrogen at the 2' position instead of a hydroxyl group
These base pairs are joined together by hydrogen bonds. The pairs themselves are able to connect based on their structures. These structures are called purines and pyrimidines.
Purines have an additional ring where pyrimidines do not. Adenine and Guanine are purines and only bond to pyrimidines
A bonds to T G bonds to C
Pyrimidines do not have the same additional ring in their structure that purines do. Thymine and cytosine are pyrimidines and only bond to purines.
One of the strands is oriented in the 3' to 5' direction, this is the leading strand. The other strand is oriented in the 5' to 3' direction, making this is the lagging strand. As a result of their different orientations, the two strands are replicated differently.
Lagging Strand
Numerous RNA primers are made by the primase enzyme and bind at various points along the lagging strand.
Chunks of DNA, called Okazaki fragments, are then added to the lagging strand also in the 5' to 3' direction. This type of replication is called discontinuous as the Okazaki fragments will need to be joined up later.
Leading Strand
A short piece of RNA called a primer, produced by an enzyme called primase, comes along and binds to the end of the leading strand. The primer acts as the starting point for DNA synthesis.
DNA polymerase binds to the leading strand and then goes along it, adding new complementary nucleotide bases to the strand of DNA in the 5' to 3' direction. This sort of replication is called continous.
Once all of the bases are matched up, an enzyme called exonuclease trips away the primers. The gaps where the primers were are then filled by more complementary nucleotides. This new strand is proofread to make sure there are no more mistakes in the new DNA sequence.
Finally, an enzyme called DNA ligase seals up the sequence of DNA into two continuous double strands. The result of DNA replication is two DNA molecules consisting of one new and one old chain of nucleotides. This is why DNA replication is described as semi-conservative, half of the chain is part of the original DNA molecule, half is brand new.
Second step of Transcription: Elongation In the process of elongation, the new RNA nucleotides are added to the 3' end of a growing chain. The new strand can ONLY be replicated in one order, from the 5' to 3' end, so the bottom template strand of 3' to 5' is chosen to make the new 5' to 3' strand of mRNA. After this is completed, the DNA strands re-form a double helix.
Third and Final step of Transcription: Termination Once the RNA polymerase reaches a transcription termination site on the DNA, the RNA transcript is released, and the polymerase detaches from the DNA. Transcription is then stopped, with the completion of a 5' to 3' end RNA strand. However, in Eukaryotes, the termination of transcription is a bit different. A sequence of "AAUAAA" is a signal for the cell to cut the newly formed strand of pre-mRNA and release it from the DNA. At the 5' end of the strand, a modified G nucelotide named CAP is added, and at the 3' end of the strand, near the AAUAAA sequence, a polyA tail is added by the enzyme polyA polymerase. (The 5' CAP will be used for translation and the 3' polyA tail helps with the stability of the mRNA.) These alterations are ONLY found in Eukaryotes. Before we can move on to translation, the pre-mRNA has to go through RNA processing before it can exit the nucleus. This process involves the removal of introns and the joining together of exons.
Translation can be defined as the formation of proteins from an mRNA template. This process consists of three phases: initiation, elongation, and termination.
The eukaryotic mRNA which is a substrate for translation consists of a unique 3' end named the poly-A tail. This mRNA also has codons that encode for specific amino acids. The 5' end of the tail is a methylated cap.
This process occurs inside a ribosome. The small ribosomal subunit binds to an mRNA cap and moves to the initiator tRNA that connects to the start codon (AUG) with its complimentary anti codon (UAC). Attached to tRNA is methionine that cooresponds to the AUG codon. The large ribosomal subunit then connects as well to complete the translation initiation complex by creating the P and A sites.
The first tRNA occupies the P site as the second tRNA enters the A site as it's complimentary to the second codon. The methionine is transferred to the A-site amino acid. The beginning tRNA detaches itself to leave the ribosome, causing a shift to create space for the next tRNA. This is the process of elongation.
The previous process of exiting tRNA's for movement of the ribosome along the mRNA to allow for the next tRNA continues until a stop codon is reached in the A-site (those being UAG, UAA, AND UGA). A release factor enters the A-site and translation is terminated.
When termination is reaches the ribosome dissociates and the newly formed protein is released
This allows for a polypeptide chain to grow until the stop codon is reached, causing the polypeptide to then float away and enter a cell organelle for folding and further modification.
In this process, the mRNA acts as a code for a certain protein. This is made possible through the bases on mRNA that are called codons which code for specific codons named anticodons (carried by the tRNA strand).
The start codon, AUG, begins translation while stop codons UAA, UAG, and UGA terminate it.