O2
FAD
NAD+
Coenzyme A
Citric Acid Cycle
- Step 1: acetyl CoA + Oxaloacetate —> Citrate
- Step 3: Isocitrate—> alpha ketoglutarate
- 1 ATP made
- 3 NADH made
- 1 FADH2 made (only made in Krebs cycle)
Also called the Krebs cycle. No enzymes are given.
FADH
Co2
Pyruvate Ozidation
Pyruvate formed in Glycolysis then enters mitochondria and is oxidized in presence of O2. The product of oxidation enters the Citric acid cycle resulting in more electron carriers NADH and FADH.
- Enters the mitochondria and is oxidized to form NADH and Acetyl Coenzyme A, which feeds into the Citric cycle. 1 pyruvate—> 1 acetyl CoA + 1 NADH
- O2 required
NADH
Acetyl COA
ADP
Substrate level phosphorylation
This is how ATP is made. An enzyme interacts with a substrate that has a phosphate group. This reaction leads the formation of a product and transfer of the phosphate group from the substrate to ADP to form ATP.
Glycolysis
Electrons are extracted from food (glucose and added to an electron carrier, NAD+. This process occurs in the cytoplasm outside the mitochondria.
Step 1: Glucose —> Glucose -6-phosphate
(hexokinase)
Step 3: Fructose —> fructose 1,6 bisphosphate
(phosphofructokinase)
Net ATP
NADH
pyruvate
Energy Investment Phase
No ATP is made through the first 5 steps of Glycolysis.
Energy Payoff Phase
2 molecules of G3P are used throughout this phase. More ATP is made in these last 6-10 stages then used in the first 5 stages. Hence receiving payoff.
Fermentation
This occurs when O2 is not present. The electron transport chain will stop working and oxidative phosphorylation will cease to occur. In the absence of O2 cells will begin to generate ATP using fermentation. This process starts with glycolysis.
Transcription
transcription factors
have two levels: general and specific. General brings low (basal) levels. Specific changes levels (increases/reduces)
promoter
RNA Polymerase (Prok)
enzyme that transcribes DNA into mRNA
RNA Polymerase II (Euk)
enzyme that transcribes DNA into pre-mRNA
use activators/inhibitors
use of activators and inhibitors to help increase or decrease level of transcription
compact gene
compact the gene so that it is not accessible to enzymes and proteins for transcription
Gene Regulation
Operons
A functioning unit of DNA containing a cluster of genes under the control of a single promoter
Lac operon
lac operon: example of both positive and negative regulation
glucose present
everything blocked, so switched off
lactose present
cAMP is on, CAP is on, Adenylyl cyclase on
structural genes
Lack: regulatory gene
LacZ: beta galactosidase
LacY: permease
LacA: trans-acetylase
Repressors
operator
Nucleosomes
DNA
Chromosomes
chromatin
fibrous double stranded DNA with protein attached
genes and proteins
Histones
H1, H2A, H2B, H3, H4
types of histones; H1: linker histone; H2A, H2B, H3, H4 make up histone core (octamer}
DNA Expression
Translation
Small Subunit
Large Subunit
tRNA
Codon Chart
Stop/Start Codon
Protein destinations
Membrane Protein
Secretion
ER
Transcription
Start Point/ Template Starnd
mRNA
For transcription in eukaryotes, it occurs in the nucleus. Transcription and Translation are not coupled, so this means that pre-mRNA is created first.
Initiation in eukaryotes is made possible by RNA polymerase II.
Transcription in prokaryotes occurs in the cytoplasm. Because transcription and translation are coupled, mRNA can be made right away.
Initiation
Initiation in prokaryotes is possible because of RNA polymerase
Elongation
Termination
Mutations
Nonsense
Missense
Silent
Frameshift
DNA double stranded
Using Chargaff's data, Roslind's X ray diffraction pattern, Watson and Crick came up with the double helix model of DNA. This would satisfy the Xray pattens where the two strands of DNA were antiparallel.
Base Pairings
Due to Rosalind and Franklins X ray diffraction image of DNA., Watson and Crick concluded that purine had to interact with pyrimidine to account for the diameter of the molecule see in the image of Rosalind and Franklins Xray. Are connected by hydrogen bonds.
Structure of a DNA strand
Made up of a sugar phosphate backbone and nitrogenous bases. Connected each nucleotide is a phosphodiester bond.
DNA Structure
Fedrick Griffith Experiment
Fedrick Griffith, a British medical officer, was developing a vaccine against pneumonia; in this process, he studied Streptococcus pneumonia. He had two strains: S strain (disease-causing) and R strain (nonpathogenic). The S strain had a smooth surface because of a capsule present, while the R strain lacked the capsule. This capsule made S strain pathogenic. When injecting the mouse with both strains, with the S strain, the rat died, and with the R strain, it lived. However, he was able to conclude that when the S strain was heated, the rat lived, but when the R strain was added with heat to the S components, it died. Seeing this, he found that something in the S strain was able to change the genetic making of R to S. Through this, they believed that it was proteins that created genetic material.
Bacteriophages
The discovery of viruses created new ideas. A bacteriophage, which is a virus, attacks bacteria. The bacteriophage is made up of proteins and DNA. This discovery narrowed down the possibility of what created the information to make viruses and limited the options of where genetic material resided.
Hershey and Chase Experiment
They proved that DNA carried genes and not proteins. How did they? The labeled DNA of one tube of bacteriophages with radioactive phosphorus and the proteins of bacteriophages in another tube with radioactive sulfur. They infected bacteria with each. They then mixed the bacteria and bacteriophages in a tube. Then, after some time, they released the bacteriophages from the bacterial surface after they injected their genes into the bacteria. They then centrifuged the bacteria cells while looking in the supernatant and pellet for radioactivity. After conducting this experiment, they concluded that they could only find radioactivity inside the bacterial cells when using radioactive phosphorus once and for all, concluding that DNA was used instead of protein.
X-ray Diffraction
Chargaff's Rule
Chargaff's rule states that for every amount of Adenine equals the amount of Thymine and the amount of Guanine equals the amount of cytosine.
Intramolecular
Ionic bonds
Non-polar Covalent
Polar Covalent
Polarity
Intermolecular
Non-Polar
Van Der Waals
Hydrophobic
Polar
Dipole-Dipole
Water properties
Solvent Polarity
High specific heat
Cohesion & Adhesion
Surface Tension
Expansion upon freezing
High heat of Vaporization
Ion-Dipole
Capsule and Slime Layers
Fimbriae
Peptidoglycan
Cytoplasm
DNA Replication
in nucleus
in cytoplasm
enzymes and proteins
DNA polymerase III
nucleotides
5' to 3' direction
Topoisomerase
overwinding
Single-stranded binding (SSB) protein
Single-stranded DNA
DNA separated
Helicase
DNA helix
Replication fork
lagging strand
discontinuously synthesized
RNA primase
Makes an RNA primer at 5' end of leading strand an dof each Okazaki fragment of lagging strand
an RNA primer
Okazaki fragments link together
DNA ligase
leading strand
synthesized continuously
DNA polymerase I
RNA primase synthesizes RNA primer
Semiconservative
one strand is conserved
Phosphodiester linkages
Bond that forms between the phosphate to link nucleotides
polyneucleotides
Repeatings units of nucleotides linked by a phosphodiester linkage.
Ribonucleic acid (RNA)
single strand
Phospholipid bilayer
Lactic Acid Fermentation
In this type of fermentation pyruvate is reduced to form lactate and recycling back NAD+ so glycolysis can continue. NO CO2 is produced.
NADH+
Lactic acid
Alcohol Fermentation
Alcohol fermentation begins when no O2 is present. Pyruvate will form acetaldehyde which then is reduced to form ethanol. CO2 is then released in this step. In the process of reduction electrons from NADH are transferred to Acetaldehyde recycling NAD+ so glycolysis can continue on.
NAD+
Ethanol
Oxidative Phosphorylation
Is the second way ATP can be made. This is where energy is used to add a Pi to ADP to form ATP. NADH and FADH2 carry electrons down the electron transport chain and end up generating ATP through this process.
Cellular Respiration
Glucose is oxidized and oxygen is reduced. Glucose with C-H bonds that have electrons equally shared is a high energy molecule. During a process involving other steps help the transfer of hydrogen to oxygen resulting in the formation of CO2 and H2O which have unequal sharing of electrons and low free energy. This energy released in this process is used to make ATP.
Aerobic respiration
Anaerobic Respiration
Photosynthesis
Calvin Cycle
needed to reduce CO2
3 Phosphoglycerate (3P)
Glyceraldehyde-3-phosphate (G3P)
Glucose
Ribulose Biphosphate (RuBP)
CO2 acceptor
Rubisco
Stroma
Chloroplasts
Chlorophyll
Light Dependent Reactions
ADP + P1
Thylakoid membranes
Photosystem 2
The reaction-center chlorophyll a absorbs at 680 nm hence called P680
The reaction-center chlorophyll
a
absorbs at 680 nm
hence called P680
Photosystem 1
The reaction-center chlorophyll a absorbs 700 nm hence called P700
Energy from sunlight
input for photosynthesis
CO2
H2O
ba
output for photosynthesis
released O2
outputs for photosynthesis
protons
electrons
NADP+
NADPH
Organic molecules+O2
Cell Respiration
ATP
Heat Energy
Stages
3. Response
Activation of Specific gene by growth hormone:
Signal Molecule enters nucleus
Activates transcription factor which binds to specific genes in DNA
Stimulates the transcription of the gene into mRNA
The mRNA is translated into a specific protein
2. Transduction
Specific ions can flow through and rapidly change the concentration of the ion inside the cell
Activated GPCR activates adenyl cyclase (enzyme)
Activated AC converts ATP to cAMP as a second messenger (amplifies signal)
cAMP converts to AMP to turn pathway off
Activates Protein Kinase 1
Protein phosphatase catalyze the removal of phosphate groups making the protein inactive again
PK1 activates PK2 by removing a phosphate group from ATP and adding it to PK2
Active PK2 phosphorylates a protein by removing a phosphate from ATP and adding it to the protein which then brings about a cellular response
Activates the G Protein Switch
Phosphotase removes a phosphate group and turns GTP into GDP
1. Reception
Ligand binds to a receptor protein
Molecule released by a cell which is received by another cell
Recepetor detects a chemical signal
Present in target cell that receives the signal molecule
Types of membrane receptors:
G Protein-coupled receptor
Ligand activates GPCR with GTP attached
Ligand-gated ion channel
A type of membrane receptor with a region that can act as a "gate" for ions, opening or closing the channel when the receptor assumes alternative shapes.
Ligand binds to receptor and opens the channel
Ligand leaves receptor and channel closes
Plasma membrane
genes
Ester linkage
Forms through a dehydration reaction.
Unsaturated Fatty Acid
These come from plant sources and are liquid at room temperature. There is one or more covalent bonds found within the carbon chain, these molecules don't have hydrogen at every position along the carbon chain. The presence of double bonds in unsaturated fatty acids can create cis/trans isomers of fatty acids.
Trans Fatty Acid
Trans= opposite. A fat molecule containing trans fatty acids are called trans fats.
Cis Fatty Acid
Cis= same. The presence of double bond in cis create a kink, a slight bend compared to the double bonds in trans fatty acids.
Saturated Fatty acid
Solid at room temperature an example of a saturated fat is butter. There are no double bond present.
Beta Glucose
Present in cellulose
Cellulose
Cellulose provides structure in plants. It is found in plant cell walls. Cellulose is made of beta-glucose. Glucose monomers are connected through 1-4 glycosidic linkages to form long chains. Glycosidic linkages that involve beta glucose molecules do not have a helical shape, chains are linear and no branching is present in cellulose.
Microfibrils
Are formed through the long chains of cellulose through the hydrogen bonds that hold the parallel chains together. Microfibrils are a strong building material for plants as well as humans. Because the human body can not digest these materials as well as plants cellulose in food passes through the digestive tract as insoluble fiber.
Alpha Glucose
Present in glycogen and starch
Dextran
Starch
Starch is made of repeating units of alpha glucose connected through 1-4 glycosidic linkages. Starch is a storage polysaccharide in plants. Starch can be digested because we have the enzymes to break them down.
Amylose
Amylose one type of starch that has no branching.
Amylopectin
Amylopectin is one of the two types of starch this starch has branching. Amylopectin has a(1,4) glycosidic linkages and the branchpoint has a(1,6) glycosidic linkages.
Glycogen
A polysaccharide formed of alpha glucose monomers connected through 1-4 glycosidic linkages. Glycogen is a storage polysaccharide in animals.
Cells
Chemical bonds
Cytoskeleton
Polypeptide Intermediate Filaments
Actin Microtubules
Tubulin Microfilaments
Vacuoles
Chloroplast
Lysosomes
Flagella
Endomembrane System
Golgi Apparatus
Endoplasmic Reticulum
Rough
Smooth
Nucleus
Cilia
Vesicles
Cell Wall
Cytoplasmic Membrane
cytoplasm allows transport, maintain cell shape and structure, protection, storage, and acts as host to metabolic processes
Saturated & Unsaturated bonds
molecules
cholesterol
Passive Diffusion
Facilitated Diffusion
Active Transport
Eukaryotes
Domain Eukarya
Plant Cells
Animal Cells
Prokaryotes
Domain Archaea
Extremophiles
organism that can survive and thrive in extreme environments (extreme temp, pressure, radiation, salinity, or pH levels)
Thermophiles
microorganisms that thrive at high temperatures; can specifically stand extreme heat
Domain Bacteria
Structure
Flagellum
Cell wall
Ribosomes
Pili
Nucleoid
Capsule
Plasma Membrane
Amino acid
Carboxyl group
Amino group
nucleotides
Phosphate
5 carbon Sugars
Ribose (RNA)
Deoxyribose (DNA)
Nitrogen Containing (nitrogenous) base
Purines
Linkage N-9 to C1'.
Guanine (G)
Adenine (A)
Pyramidines
linkage N-1 to C1'.
Uracil (U)
Thymine (T)
Cytosine (C)
TRNA
Brings amino acids to the ribosome during the synthesis of a polypeptide
Deoxyriboneucleic acid (DNA)
Provides directions for its own replication. Directs synthesis of messenger RNA and controls protein synthesis, a process called gene expression. Double Helix
Quaternary structure
Two or more polypeptides joined together
Hydrogen Bonds
Occurs between H and O-s in R groups
Disulfide bridges
Occurs between S elements
covalent bond
Ionic bond
Occurs between basic and acidic R groups
Hydrogen bonds
Beta pleated sheet
hydrogen bond between two or more parallel structures
Alpha helix
hydrogen bond between every 4th amino acid
Secondary structure
Tertiary structure
3-dimensional shape stabilized by interactions between side chains
Primary structure
linear chain of amino acids
Basic R group
positive charge
Acidic R group
negative charges
Polar side chain
Hydrophillic. Includes OH, NH, SH, and CO compounds.
Nonpolar side chain
hydrophobic. Includes CH molecules
Side chain (R group)
Peptide bonds
bond between amino acids
Polypeptide
A polymer of many amino acids linked by peptide bonds
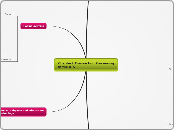
Nucleic Acids
Lipids
Phospholipids
Phospholipids are a type of lipid. There is one glycerol linked to two fatty acids. At the third OH in glycerol another group is attached that contains a phosphate group. They are amphipathic.
Hydrophobic tail
Hydrophilic head
Sterols
A type of lipid. Contain 4 fused rings.
Cholesterol
An example of a steroid is cholesterol. This is found in animals and a common component of membranes.
Found embedded within the membrane. Helps with membrane stability.
HDL
Good Cholesterol. Good to have.
LDL
Bad Cholesterol. Hydrogenation can cause some trans fats to form. Trans fats can increase LDL levels. Having high LDL and low HDL is bad to have.
Fats (triglycerides)
Glycerol
Fatty acid
Carbohydrates
Monosaccharides
Combine to form different types of polymers called polysaccharides. The simplest sugars are called monosaccharides, made up of C, H, OH, and CO groups.
Polysaccharides
These are formed when 100 or more monosaccharides are bonded together through glycosidic linkages. There are two basic functions of polysaccharides in cells that serve different functions- storage and structure. The structure and function or a polysaccharide are determined by its sugar monomers and the types of their glycosidic linkages.
Structural Polysaccharides
Organisms build strong materials from this.
Storage Polysaccharides
Plants and animals store sugars for later use.
Disaccharides
Is formed when dehydration reaction joins two monosaccharides. This covalent bond created is called a glycosidic bond/linkage.
Glycosidic linkages
The glycosidic linkage formed is named based on which carbon atoms are involved in the formation. So if 1 and 2 monosaccharides are involved the bond is called 1-2 glycosidic linkage.
Proteins
Constructed from the same set of 20 amino acids, linked in unbranched polymers. Made up of 1 or more polypeptides folded and coiled into 3D structures
Cell signaling
Long distance
Endocrine (hormonal) signaling
Specialized endocrine cells that secrete hormones into the body fluid
Local
Synaptic
A nerve cell releases neurotransmitter molecules into a synapse, stimulating the target cell (ex. muscle)
Paracrine
A signaling cell acts on nearby target cells by secreting molecules of a local regulator (ex. growth factor)
from glycolysis
Electron Transport Chain
The ETC is located in the inner mitochondrial membrane. Complexes 1, II, III, iV, Q and Cyt c all make up the ETC. Complexes 1,2,3, and 4 are electron pumps. The more protons the lower the pH. Complexes 1, III, IV are also H+ pumps. Their job is to pump H+ against conc gradient in the intermembrane space. When electrons are transferred down ETC energy is released. This released energy is used to pump H+ against the concentration gradient. NADH transfers electrons to complex I, while FADH2 transfers electrons to complex II. Electrons end up ultimately in oxygen which form water.
Chemiosmosis
H+ in the intermembrane space go back down their concentration gradient through an ATP synthase. Proton motive force is then used to add Pi to ADP to form ATP.
Macromolecules