Floating topic
Diffuse directly across the lipid bilayer of the plasma membrane to encounter its receptor in the cytoplasm
Color Code:
DNA Structure, Replication, Expression and Regulation
Energy and cell communication
Cell structures
Cell structures that all cells share
Group:
Shalimar Juarez
Layla Williams
Samantha Gobert
Kayla Tran
William Knudsen
Animal Cells
Plasma Membrane
Membrane Receptor
Ion channel receptor
On the post synaptic cell membrane
Channel remains closed until ligand binds to the receptor
Channel opens but only a specific ion is able to flow through the channel.
Channel closes and ions are no longer able to flow.
Tyrosine kinase receptor
Two receptor tyrosine kinase proteins form a dimer after activation
Unphosphorylated dimer --> Phosphorylated dimer
Active relay proteins produce cellular response
Each polypeptide are kinases: it takes phosphate from ATP and adds to Tyt; takes phosphate from ATP and adds to polypeptide at Tyrosine. When all 6 Tyrosine gets phosphate groups. It is now fully activated receptor. We call it Phosphorylated because it has phosphate groups added. Once fully activated, rely protein binds and cell response
Protein kinases
Enzymes that catalyze the transfer of phosphate groups from ATP to proteins
G-Protein linked receptor
Phosphatase; it can remove a phosphate group
G-protein couple receptor changed shape and binds to and activates G-protein by changing it's shape
G-protein leaves receptor to bind to and activates an enzyme called Adenylyl Cyclase
removes Phosphate and convert back to GDP
inactive
Signal Molecule binds to receptor again
Adenylyl Cyclase converts ATP to cAMP
cAMP
One Phosphate is used to make cAMP
c
Triggers a phosphorylation cascade
cAMP is going to bind to a kinase (1,2,3)
1st kinase is activated, gets to rely molecules and activate it; it gets a kinase so it will take a phosphate group and add to the next protein that is also a kinase. Now its active; it will take a phosphate group from ATP and add to 3rd kinase; and repeats; hence why it is called a Phosphorylation Cascade
As each kinase is activating the next, they themselves will be switched off due to cost of phosphate group due to phosphatase
To amplify the signal
Activates transcription factor for gene expression
phospholipids
Help Cell membranes and membranes surrounding organelles to be flexible and allows for vesicle formation and enables substances to enter or exit a cell through bulk transport
Lipids
nonpolar
molecules
hydrophobic
interactions
nervous system
myelin sheaths
Fatty acids
Unsaturated
Short Tail length
Membrane Fluidity
Double covalent bonds
Plants
Saturated
Animals
chemical messengers,
energy storage
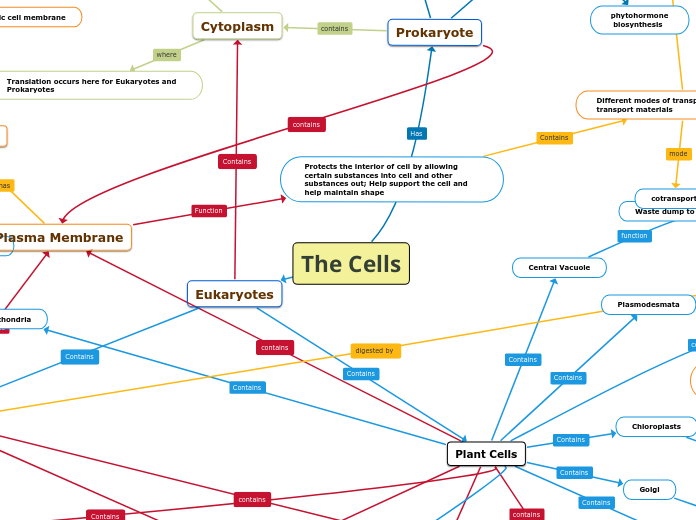
Protects the interior of cell by allowing certain substances into cell and other substances out; Help support the cell and help maintain shape
Different modes of transport used to transport materials
cotransport
occurs when active transport of a solute indirectly drives transport of other substances
Facilitator
Allow diffusion process to take place in membranes that are made up of glycoprotein
Passive Transport
osmosis
diffusion of free water across a selectively permeable membrane
diffusion
tendency for molecules of any substance to spread out evenly into available space as a result of thermal motion
No energy required, moves from high to low concentration
Active Transport
Ions moving across
their concentration gradient
Requires energy in the form of ATP. Moves from a low to high concentration to transport through cell membrane
Bulk
Exocytosis - contents
leaving the cell
Move very large molecules across a membrane using vesicles
Proteins and Carbohydrates
Endocytosis-taking
something into cell
Receptor-mediated
endocytosis
specialized type of pinocytosis that enables the cell to acquire bulk quantities of specific substances
Pinocytosis
when the cell takes in extracellular fluid from outside in vesicles
Phagocytosis
when a cell engulfs large food particles/other cells by extending part of its membrane out
Secondary Active Transport
Transport of small molecules that uses an established electrochemical gradient to power the movement
r
Transport of small molecules that directly uses ATP as an energy source
Cellular Respiration
Pathway that releases energy by breaking complex molecules into simpler compounds
C6H12O6 + 6O2 → 6CO2 + 6H2O
Step 1: Glycolysis
Fermentation
Alcohol Fermentation
To produce ATP under anaerobic conditions
Lactic Acid Fermentation
Glucose is converted into cellular energy and the metabolite lactate
To convert NADH, a chemical compound found in all living cells, back into the coenzyme NAD+ so that it can be used again
Pathway that converts glucose, into pyruvic acid
Step 2: Pyruvate Oxidation and
Citric Acid Cycle
Step 3: Oxidative Phosphorylation
Electron Transport Chain
inner mitochondrial membrane
e- down ETC to O2
to form H2O in animal
cells
NADH & FADH2
Complexes I, II, III, IV
Coenzyme Q and cytochrome proteins
Chemiosmosis
photophosphorylation
when H+ go back down conc. gradient
through ATP synthase
Mitochondrial matrix
in animal cells
Stroma in plant cells
convert ADP to ATP
organization of the thylakoid membrane
PS2
photon of light is absorbed by chlorophyll, causes e- to jump to excited state, go back to ground state and releases the absorbed energy
components of the electron transport chain
PS1
photon of light absorbed by one of the pigment molecules causes e- to get excited, as they go to ground state energy is released which eventually reaches the main chlorophyll molecules
Electrons are exchanged between molecules, which creates a chemical gradient that allows for the production of ATP
26 or 28 ATP
Carries on from step one and fuels the production of ATP in the process
2 ATP
Sodium Potassium Pump
maintains the internal concentration of potassium ions [K+] higher than that in the surrounding medium concentration of sodium ions [Na+]
Used in the cellular membranes
Can take place anywhere in the cell
cell junctions
gap junctions
Allows for movement between cells
desmosomes
form stable adhesive junctions b/t cells
tight junctions
Prevents movements
Lysosomes
Breaks down biomolecules
Centrosome
Regulates cell motility
cytoskeleton
Intermediate filaments
Anchors nucleus,
forms nuclear lamina
Microtubules
Cell motility
chromosome movement,
organelle movement
Microfilaments
ER
Rough
produce proteins
Smooth
stores and synthesizes lipids
The Cells
Prokaryote
Cytoplasm
Translation occurs here for Eukaryotes and Prokaryotes
Transcription occurs in prokaryotes
5' cap and poly a tail, and introns
Step 1. Initiation
After RNA polymerase binds to the promoter, the DNA strand unwinds, and the polymerase initiates RNA synthesis at the start point on the template strand
Step 2. Elongation
The polymerase moves downstream, unwinding the DNA and elongating the RNA transcript 5' to 3'. in the wake of transcription, the DNA strands re-form double helix
Step 3. Termination
Eventually the RNA transcript is released, and the polymerase detaches from the DNA
RNA Polymerase (RNAP)
separate the 2 strands
template strand to start RNA transcription in 5' to 3' direction
starts transcription
mRNA is translated to protein
DNA to pick the template strand (the parent strand)
Daughter strands
Archaea
phytohormone
biosynthesis
Nucleoid
Area of cell that holds genetic material.
Bacteria
Fimbriae
Attachment structure on some prokaryotes
Flagella
Used for cell movement in some prokaryotes
Break down nutrients
Protects the cell from lysis and gives structure
Petidoglycan
Protects bacterial cells from environmental stress
Eukaryotes
Plant Cells
Cell Wall
Cellulose
Carbohydrate
Storage
Starch
Glycogen
Structure
Chitin
Monosaccharides
Glycosidic linkages
Covalent bond
Maintains cell shape
Central Vacuole
Waste dump to keep the cell in shape
Peroxisome
helps with oxidation reactions
metabolism, detoxification, and signaling
Chloroplasts
cells of the mesophyll, interior tissue of the leaf
30-40 chloroplasts
site of photosynthesis
Pathway that consumes energy to build complex molecules from simpler ones
to convert radiant energy from the sun into chemical energy that can be used for food
6CO2+6H2O+light energy = C6H12O6+6O2
Converts energy of sunlight to chemical energy stored in sugar molecules
the calvin cycle produces sugar from CO2 with the hep of NADPH and ATP produced by the light reactions
Plasmodesmata
facilitate exchange of signaling molecules b/t neighboring cells
Mitochondria
Generates energy in form of ATP
kinetic
associated with motion of molecules or objects
potential
stored energy
Golgi
Secrete cell productions (proteins and lipids)
Ribosome
translation occurs
mRNA and initiator tRNA
adheres to start codon
to complete translation initiation complex
enters ribosome and carries an amino acid
covalently bound to Met
using peptidyl transferase
leaves ribosome, and shifts over for the next tRNA
to the 1st two and this process continues all the way down mRNA
polypeptide chain will grow
recognize signal peptide
and transport complex to ER
where synthesis continues
cleaved by signal peptidase
stop codon is reached
the completed polypeptide swims away
mitochondria, chloroplasts,
peroxisomes, or nucleus
folding and modification
golgi through vesicles
rough ER or lysosomes
plasma membrane
Proteins
quaternary
Denaturation
interchain interactions
Noncovalent bonds between complementary surface hydrophobic and hydrophilic regions on polypeptide subunits
tertiary
polypeptide folds through R groups to form three dimensional shape
Depends on the R group
Different types of R groups
Basic
pH > 7, positively charged
Acidic
pH < 7, negatively charged
Polar
Outside of membrane or surface
Hydrophilic
Having a tendency to mix with, dissolve in, or be wetted by water
Electrons are shared
unevenly btwn atoms
Having OH, SH, or NH groups
Non-Polar
Inside of membrane or not on surface
Hydrophobic
Electrons are
shared evenly
between atoms
Having H or CH or Carbon rings
Polar and Nonpolar are going to form hydrophobic and hydrophilic bonds
Acidic and Basic R groups are going to form Ionic Bonds
secondary
alpha helices, and beta pleated sheets
Hydrogen Bonds
Bonds between polar covalent
molecules that contain hydrogen
attached to F,O, or N
primary
amino end and carboxyl end
Peptide Bonds
Nucleus
Intracellular Receptors
Steroid Hormones
binding of hormone w/ receptor causes receptor to be activated and changes its shape
through nuclear pores, binds the DNA and RNA is made through transcription, proteins are made through translation
by making proteins
because a signal molecule came in contact with the receptor
The Steroid hormone aldosterone passes through the plasma membrane
Aldosterone binds to a receptor protein in the cytoplasm activating it
The hormone receptor complex enters the nucleus and binds to specific genes
l
The mRNA is translated into a specific protein
Receptors is inside the cell
DNA
DNA Replication
Helicase separates
ds DNA into 2 parent strands,
forming a replication bubble
at ORI
1 ORI
makes RNA primers
complementary to parent strand
to add nucleotide monomers to
3' end of primer through phosphodiester bonds
Lagging strand that
requires multiple primers
Okazaki fragments
Leading strand that
requires 1 primer
primers removed and replaced
with DNA by DNA polymerase I
ligase
Mitosis
Prophase
Metaphase
Anaphase
Telophase
End of mitosis
Nuclear membranes reform
Forms two identical
daughter cells
Chromosomes condense and
divide the cell
Separates the cells into
two identical daughter cells
Somatic cells
multiple ORIs
SSB
Topoisomerase
Transfer their DNA into bacteria cells. This reprograms the cell to produce more bacteriophages.
The Hershey-Chase Experiments
Two batches of bacteriophages were grown. One with 32S(associated with proteins) and the other with 32P(associated with DNA). Each were then allowed to infect bacteria cells and were put in a centrifuge.
The pellet produced from both were examined and the 32S was found in the liquid, while 32P was found in the pellet. This concludes that the genetic material transferred is in the DNA.
transcription
A Eukaryotic promoter commonly includes a TATA box about 25 nucleotides upstream from the transcriptional start point
Several transcription factors, one recognizing the TATA box, must bind to the RNA before RNA polymerase II can bind in the correct position and orientation
bind to enhancer
sequences
bind to proximal
control elements
regulate eukaryotic
gene expression
Additional transcription factors bind to the DNA along with RNA polymerase II, forming the transcription initiation complex.
RNA Polymerase II then unwinds the DNA double helix and the RNA synthesis begins at the start point on them template
5' cap is made due to guanine nucleotide w/ 3 phosphate and a poly A tail. It is added as a protector
Pre-mRNA and mRNA
AAUAAA, is called the poly-A site
When the Pre-mRNA is made, the AAUAAA is present in an enzyme called Ribonuclease. It comes along and breaks the phosphodiester bond, releasing the pre-mRNA .
an enzyme called poly A polymerase will add 100-200 As at the 3' end
RNA Processing
introns, exons, 5' cap, 3' polyA tail
exons, 5' cap and 3' poly A tail
RNA splicing; the process of removing introns
through Spliceosome; is a complex of proteins and RNA that does the splitting
to make different proteins called Alternate slicing
RNA Polymerase II
promoter
the 2 strands
template strand and start making RNA transcription in 5' to 3' direction
pre-mRNA
mRNA is produced by RNA polymerase II with the help of transcription factors and binds to promoter
nontemplate
template strand
mRNA
at the start codon, moves downstream
from 3" to 5", and generating mRNA from 5' to 3'
leaves nucleus
Translation in cytoplasm
mRNA is translated to form proteins
Tested if DNA can transfer between bacterias by injecting them into a mouse and seeing if it survived or not. Mixed heat killed smooth pathogenic cells with rough nonpathogenic cells.
Resulted in heat killed smooth cells transferring its DNA to the rough cells, turning them into smooth cells.
Nucleic acid
Nucleotides
sets of 3 are used to lay down amino acids to form a polypeptide and that secret code is the Codon Chart
Phosphate
Nitrogenous base
DNA from any species of any organism should have a 1:1 stoichiometric ratio of purine and pyrimidine bases
A = T and G = C
Pentose
phosphodiester linkages
Basis of activity and growth
genes, and structures that have hereditary information