Glycosylation: Attaches carbohydrates to organic molecules such as proteins.
Free Ribosome: Ribosome that is not attached to any cellular structures. Found in the cytoplasm.
Stop Codon: AG, UGA, or
UAA.
Signal Recognition Particle (SRP): Ensures proteins are correctly targeted to the ER membrane.
Start Codon: AUG
3 prime end
Poly-A Tail
5 prime end
Terminator Sequence: Nucleid acid sequence that indicates the end of transcription.
Premature mRNA: Primary transcript.
Mature RNA (mRNA): Carries genetic information.
GPPP Cap
Template Strand (TAC): Used to build the RNA. 5' to 3' direction.
Exons: Contain coding information for protein.
DNA Ligase: Connects DNA together.
DNA Polymerase III: Needs a primer and only adds 5’ to 3’ to the daughter strand. Builds the
DNA strand.
DNA Polymerase I: Needs a primer and only adds 5’ to 3’ to the daughter strand. Replaces RNA
with DNA.
Single Stranded DNA Binding Protein (SSB): Keeps the parent strands separate.
Topoisomerase: Keeps the strands from tangling. Relieves tension in the parent strand from
overwinding.
Transcription Factor: Controls gene expression.
Promoter Sequence: DNA sequence that defines where transcription begins.
RNA Polymerase II: Transcribes protein-coding genes to create mRNAs. (Eukaryotes)
RNA Polymerase: Transcribes DNA to RNA. (Prokaryotes)
Release Factor: Ends translation by recognizing a stop codon in the mRNA.
Newly Formed Polypeptide
Large Ribosome Subunit
A Site: First place tRNA attaches.
P Site: Holds the tRNA which is linked to the growing polypeptide chain
Peptide Bond
E Site: Releases tRNA.
Small Ribosome Subunit
Peptidyl Transferase: Catalyzes the formation of peptide bonds between amino acids
Transfer RNA (tRNA): Serves as the link between mRNA and the amino acid chain.
Poly-A Polymerase: Adds a poly-A tail to the 3' end of RNA molecules.
Spliceosome: Removes introns from pre mRNA.
Introns: Non-coding sequences found in DNA or RNA. Prokaryotes do not carry introns.
Protein Kinase A (PKA): Enzymes that catalyze the transfer of phosphate groups from ATP to proteins
Ribonucleotides: Forms the backbone of RNA molecules through phosphodiester bonds.
Floating topic
Topic 1
Topic 2
Photosynthesis
Calvin Cycle
-The Calvin cycle produced sugar from CO2 with the help of the NADPH and ATP produced by the light reactions
-CO2 is initially incorporated into an organic molecule through a process called carbon fixation
-ATP provides the necessary chemical energy and NADPH provides electrons needed to reduced CO2
3 CO2 enter and there are 3 Ribulose Bisphospate. Rubisco takes the CO2 and RuBP and makes a 6 carbon molecules that is unstable which then becomes two 3 Phosphoglycerate, for a total of six 3 phospholyerate. 6 NADPH and 9 ATP get inputted to the cycle and as a result 1 G3P is made from the whole cycle.
Light Dependent Reactions
-The light reactions convert solar energy into chemical energy
-H2O is split to provide electron and protons
-Oxygen is released as a waster product
-The electron acceptor NADP+ is reduced to NADPH
- ATP is generated by adding a phosphate group to ADP in a process called photophosphorlyation
Light energy from the Sun excites electrons in chlorophyll molecules of PSII. The electrons are replenished by splitting water, which produces oxygen, protons and electrons. Then the excited electrons are passed through an electron transport chain where their energy is used to pump portions into the thylakoid lumen. Electrons are now at PSI and light re-energiezes these electrons in PSI. Now these electrons reduces NADP+ to NADPH. Also some ATP is made because of the electron transport chain
Topic 3
RNA Structure
-Monomer: RNA is also made of nucleotides, which include:
A ribose sugar (a 5-carbon sugar with a hydroxyl group at the 2' position), a phosphate group, a nitrogenous base (adenine, uracil, guanine, or cytosine).
- Structure: RNA is usually single-stranded and shorter than DNA. It can fold into complex secondary structures due to base pairing (e.g., hairpins). In RNA:
Uracil (U) replaces thymine (T) and pairs with adenine (A).
- Bond Between Monomers: Similar to DNA, RNA nucleotides are joined by phosphodiester bonds.
DNA Structure
- Monomer: DNA is composed of nucleotides, which include:
A deoxyribose sugar (a 5-carbon sugar lacking one oxygen atom at the 2' position compared to ribose in RNA), a phosphate group, and a nitrogenous base (adenine, thymine, guanine, or cytosine).
- Structure: DNA is a double-stranded helix. The two strands are antiparallel and held together by hydrogen bonds between complementary base pairs:
Adenine (A) pairs with thymine (T) which have two hydrogen bonds.
Guanine (G) pairs with cytosine (C) which have three hydrogen bonds.
- Bond Between Monomers: Nucleotides in a single strand are connected by phosphodiester bonds between the 3'-hydroxyl group of one nucleotide and the 5'-phosphate group of the next.
Protein Modification
ER Lumen: Where post-translation modifications occur. Either folds or refolds the polypeptide goes through glycosylation or packages proteins into vesicles.
Glycoprotein: Protein that goes through glycosylation. Aids the immune system.
Transport Vesicle: Carries proteins to different organelles.
Golgi Apparatus: Packages the proteins to transport them to their destinations.
Endoplasmic Reticulum: Proteins return for further modifications.
Lysosome: Proteins are broken down and recycled.
Plasma Membrane: Regulates the transport of materials entering and exiting the cell.
Gene Regulation
Nucleosome
DNA
wraps around histones, linker DNA used to 'link' it all together
Histones
Histone Core/Octomer
two of H2A, H2B, H3, and H4
Types: H1, H2A, H2B, H3, H4
H1 is only used to help package DNA
Eukaryotic
DNA Bending Protein
bends to aid in transcription
TATA box
shows RNA Poly where to bind
Cell Specific Transcription
enhancers and activator proteins
Gene Expression
High Levels
Gene is Expressed
Basal
low or background expression, not very present
Gene regulation occurs in transcription
Distal Control Elements
Binds to specific transcription factors (activators/repressors)
Enhancers- sequences in DNA upstream or downstreamof gene
Proximal Control Elements
sequences in DNA close to promoter and binds to general transcription factors
Prokaryotic
Negative Regulation
Presence of a repressor to inhibit gene expression, lack of one means gene will be expressed
Positive Regulation
Presence of an activator binding to the operator in order to express a gene, lack of activator means gene will not be expressed
Operons
lac operon
Is it on or off?
Is Lactose Present?
Is Glucose Present?
Lactose
Adenyl Cyclase Active
cAMP levels High
Operon is ON
Adenyl Cyclase Inactive
cAMP levels low
Operon is OFF
Repressor bound to Operator
OFF
Regulatory Gene
Lac I
repressor protein
Always on
Lactose utilization in E coli.
both negative and positive regulation
Lac A
Transacetylase
Lac Z
Beta Galactosidase
Lac Y
Permease
Structural Genes
multiple genes
Operator
Repressors and activators bind here
The 'switch' located near or in the promoter
Promoter
bound by RNA poly II to start transcription
a cluster of functionally related genes that can be under coordinated control of a single on/off switch
DNA Replication
Parental DNA: Serves as the template.
Origin of Replication: Site where DNA replication starts.
Helicase: Separates the parent strand into two individual strands.
Replication Bubble: Open region of DNA after helicase unwinds seperates the strands.
Replication Fork: Initiates replication.
DNA Primase: Makes up the RNA primer.
DNA Primer: Starting point for DNA synthesis.
Lagging Strand (3' to 5'): Moves against the fork. Replicated inconsistently in small fragments.
Okazaki Fragments: Short DNA sequences that are synthesized discontinously.
Leading Strand (5' to 3'): Moves towards the fork. Consistently synthesized.
DNA Translation
Termination: The ribosome encounters a stop codon in the A site, which signals the release factor to let go of the newly formed polypeptide.
Elongation: Ribosome moves along the strand and starts to attach amino acids. An amino acid chain is formed.
Initiation: A small ribosome subunit binds to the mRNA 5' end, scans for the start codon, and starts translating.
DNA Transcription
Termination: Ends trancription process by marking the end of the gene.
Elongation: RNA Polymerase moves along the strand, adding nucleotides and rewinding the strand.
Initiation: RNA Polymerase binds to promoter and unwinds DNA helix to access the template strand.
Cellular Respiration
-A metabolic pathway that uses glucose to produce adenosine triphosphate (ATP )
Glycolysis
-happens in the cytoplasm/ can happen without O2
1.One glucose molecules go in and gets turned into Glucose 6 Phosphate with the enzyme hexokinase (1 ATP need for this/ the ATP becomes ADP)
2. The glucose 6 phosphate magically becomes fructose 6 phosphate which gets turned into Fructose 1 6 Bisphosphate with the enzyme phospofructokinase ( 1 ATP Needed for this/ the ATP becomes ADP)
3. End result: 2 NADH, 2 Pyruvate, 4 ATP/ Net: 2 bc you used 2
Pyruvate Oxidation
-Happens in the Matrix
CoA gets attached to 1 Pyruvate and that makes 1 Acytyl CoA this process turned NAD+ to NADH
Citric Acid Cycle
- Happens in the matrix also know as the kreb cycle
-Oxaloacetate reacts with acetyl-CoA to produce citrate after a lot of stuff the citrate becomes isocitrate which then becomes ketogluterate and a NADH is produced the last steps of this cycle produce 1 ATP, 2 NADH and 1 FADH2
- Total Products for 1 cycle: 3 NADH (which are electron carriers), 1 ATP, 1 FADH2 ( which are also electron carriers)
Oxidative Phosphorylation
Because there is a lot of hydrogen ions in the intermembrane space, they will move down the concentration gradient to the matrix. This movement down the concentration gradient through ATP synthesis gives ATP synthase to convert ADP to ATP by adding a phosphate
First electrons are given to complex 1 by electrons carriers which is NADH. When the electrons are given, NADH is oxidized to NAD+. The electron moves down from complex 1 to complex Q then to complex 3 then to cytc C then to complex 4. Complex 1 3 and 4 are proton pumps and when electrons pass through them they get activated and they push Hydrogen ions from the Matrix to the intermembrane space. Oxygen is the final electron acceptor
Energy Transfers and Transformations
Catabolic and Anabolic
Coupling
add together exergonic and endergonic, and delta G must equal a negative number for the reaction to happen
Anabolic
Photosynthesis
Consumes energy to build longer and more complicated molecules from simpler ones
Endergonic
Catabolic
Cellular Respiration
energy is released by a breakdown of a complex molecule into simpler components
Exergonic
Enzymes
Feedback Inhibition
the end product becomes an inhibitor and binds + changes the shape of the original enzyme's active site
end product molecule unbinds and the synthesis of the end product can continue.
Allosteric Regulation
Cooperativity
when one substrate molecule binds to the active site of a subunit and locks the other subunits into the active form.
Four subunits with an active site on each and a space inbetween the subunits to bind the regulatory molecules
activators and inhibitors
Inhibitors stabilize the enzyme but inhibit the function of the enzyme and does not allow the binding of other substrates
Activators stabilize the enzyme into a Stabilized Active Form
Functions
Noncompetitive Inhibition
function is only carried out to an extent or not at all
substrate may still be able to bind
particle binds to enzyme not at the active site and changes the shape.
Competitive Inhibition
function is not carried out
a mimic of the substrate binds to the active site
substrate binds to enzyme's site
effected by:
pH
ex: intestinal enzyme is at higher pHs than a stomach enzyme (more acidic)
Heat
heat loving enzymes ensure the reaction can happen at high temperatures
Lower activation energies
metabolic pathways
Gibb's Free Energy
Spontaneity
delta G is positive
Delta G is Negative
more stable + released free energy can be harnessed to do work
delta G = delta H - T delta S
The lower the free energy, the more stable and less work capacity
The higher the free energy,the less stable and greater work capacity
Cell Membranes
Endocytosis
Receptor Mediated Endocytosis
takes in specific molecules in bulk once specific substance is attached to receptor
Pinocytosis
cell drinking, pinches off into vesicle
Phagocytosis
Takes in food in food Vacuole
Neurons
Action Potential
Resting Phase
Depolarization
Rising Phase
Falling Phase
Hyperpolarization
K+ channels stay open, more negative inside cell
Na+ channels close, K+ channels open and K+ leaves
All Na+ channels open, all Na+ goes inside, inside of cell is positive, outside in negative
some Na+ channels open, becomes less negative inside
below threshold, no movement, K+ and Na+ channels closed
Membrane Potential
Inside neuron is slightly negative, outside is slightly positive
Semi-Permeable
Cholesterol
above and below phase temp
maintains fluidity (not too fluid, not to rigid)
hydrophilic head, hydrophobic tail
made with fatty acids
Unsaturated
makes membrane more fluid
above phase temp
Saturated
Membrane is more solid
below phase temp
Transport
Active Transport
Sodium Potassium Pump
charges, inside negative, outside cell is positive
2 K+ into the cell, and 3 Na+ out of the cell
moves substance from Low to High; goes against gradient
DO NOT require ATP
Passive Transport
diffusion of substance across cell membrane, concentration from High to Low, no transport protein needed.
Osmosis
Animal Cell
Normal
no net movement of water
Lysed
cell blows open
Shriveled
Plant Cell
Flaccid
plant cell is limp, still has water inside, no net movement of water
Turgid
Cell is full, has a little belly
Plasmolyzed
shriveled
Toncity
Hypotonic
out to higher solute concentration
solute is higher outside the cell than inside
Hypertonic
inside, to higher concentration
solute concentration is higher inside then outside
Isotonic
in and out of cell, no net movement
solute concentration is equal inside and outside
a solute cannot move across cell membrane, so water moves across membrane to balance concentration of solute on both sides
Facilitated Diffusion
Subtopic
transport protein
Cell Signaling
Second Messengers
Cyclic Adenosine Monophosphate (cAMP): Used for intracellular signal transduction.
Phosphodiasterase: An enzyme that breaks phosphodiester bonds such as in cAMP.
Stages
Reception: Signal molecule binds to the receptor.
Transduction: Triggers a process where a molecule changes the next molecule in the pathway.
Response: Activation of cellular response.
Signals: Molecules released by a cell which is received by another cell.
Long Distance: Reaches target cells located far away in the body using a system like the bloodstream.
Hormonal Signaling: Chemical signals released into the bloodstream to reach distant locations.
Local Distance: Reaches neighboring target cells.
Paracrine Signaling: Signal that induces change to nearby cells.
Synaptic Signaling: Signal that allows neurons to communicate through the synapse to inhibit or activate the target cell>
Receptors: Target cells that receive the signal molecule.
Intracellular Receptors
Nucleus
Cytoplasm
Adenylyl Cyclase: An enzyme that converts ATP into cAMP.
G-protein: Molecular switch. Transfers signals into the cell.
Guanosine Triphosphate (GTP): Active state
Guanosine Diphosphate (GDP): Inactive state
Membrane Receptors: Signal molecules are hydrophilic.
Ion Channel Receptor: Allows specific ions to flow through the membrane in response to a signal
G-protein Coupled Receptor (GPCR): Transmembrane protein.
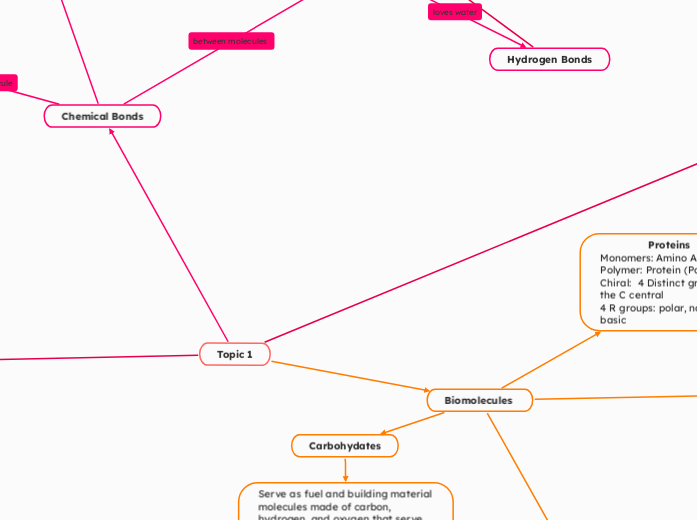
Chemical Bonds
Properties of Water
Universal Solvent
High Heat of Vaporization
water needs a high temperature to change into its gas form
H-Bonds absorb heat and then break, turning liquid into gas
Denser as a Liquid
than a Solid
ice floats, allowing life under ice layer
Expansion Upon Freezing
Ice
Insulator
crystal structure w/ stable hydrogen bonds
Cohesive Behavior
tendency for water to stick to itself, mainly caused by H-Bonds
forms H-bonds with each other, also get high surface tension
High Specific Heat
A high heat needed to raise one gram of water by one degree
H-Bonds absorb heat
Reform H-bonds and release heat
Intramolecular
Ionic
Covalent
Nonpolar
in water, get caged together
Polar
in water, form hydrogen bonds and dissolve
Intermolecular
Amphipathic
Phospholipid Bilayer
Hydrophilic Head
Hydrophobic Tail
Hydrogen Bonds
Hydrophobic
nonpolar molecules
Ion Dipole
polar and ion
positive attracted to negative charges
Dipole-Dipole
Attracted to opposite charges
Biomolecules
Carbohydates
Serve as fuel and building material
molecules made of carbon, hydrogen, and oxygen that serve as an energy source and structural component in organisms
Glycogen
-Unlike beta glucose, which has its -OH group attached above the ring, alpha glucose has its -OH group linked below the ring.
Starch
Dextran
Cellulose
Chitin
Nucleic Acids
Monomer: Nucleotide
2 type of nucleic aids: DNA and RNA
The phosphodiester bond is a covalent linkage between the phosphate of one nucleotide and the hydroxyl (OH) group attached to the 3′ carbon of the deoxyribose sugar in an adjacent nucleotide, forming what is known as the “sugar-phosphate backbone” of DNA.
DNA
Deoxyribonucleic Acid (Carbon 2 has one less O compared to RNA)
Has a double helix: antiparallel
The two strands are joined together by hydrogen bonds
Nucleotide
1. 5 carbon sugar (Pento)
2. Nitrogenous base
DNA - T, A, C, G
RNA - U, A, C, G
Purines: A, G Ag (Silver is pur=e)
Pyrimidines: U, C, T
3. Phospate Group
RNA
-Ribonucleic Acid
-Mostly exists in the single-stranded form, but there are special RNA viruses that are double-stranded.
-Type of RNA: messenger RNA (mRNA), transfer RNA (tRNA), and ribosomal RNA (rRNA)
Proteins
Monomers: Amino Acids
Polymer: Protein (Polypeptide)
Chiral: 4 Distinct groups around the C central
4 R groups: polar, non polar, acidic, basic
Lipids
Triglyceride (TAG)
Made up of 1 glycerol Head + 3 Fatty Acids
Type of Bond between FA and glycerol head: Ester Linkages
Non-polar molecule
1 TAG = 3 FA = 3 ester bonds
2 different types of FA
Unsaturated Fat
Liquid at room temp
Has double bonds
Hydrogenation is a process that can change the Unsaturated FA to Saturated by changing the double bonds to single bonds
Example: Oil
Trans Unsaturated Fat
The hydrogens are on opposite sides
Trans=opposite
Cis Unsaturated FA
The hydrogens are on the same side
cis= sisters or same side
Saturated FA
No double bonds
Solid at room temp
Example: Butter
Phospholipids
1 glycerol + 2 FA + 1 phosphate group
Is an amphipathic molecule (has a hydrophobic and hydrophilic part)
The Tails
Made up of 2 FA's
It is Hydrophobic which means it is water hating therefore it is non-polar
The Head
Made up of Glycerol and phosphate
Is Hydrophilic so it is water loving and polar
Sterols
4 fused rings
AKA steroids
Cholesterol
Found imbedded within the cell membrane
Helps with membrane stability and fluidity
High HDL and Low LDL = good to have
Low HDL and High LDL = bad to have
Cell Structures
Prokaryotes
Archaea
Bacteria
Pili: Aids in DNA transfer between bacteria.
Fimbriae: helps bacteria stick to a surface.
Flagellum: Enables movement.
Capsule: Protective layer. Helps bacteria adhere to surfaces.
Cell Wall: Maintains cell shape.
Peptidoglycan: Structural support.
Nucleoid: Contains genetic material.
Eukaryotes
Plant
Microfilament: Maintains cell shape and movement. Made of actin protein.
Vacuole: Stores water, nutrients, and waste products within the cell.
Plasmodesmata: Connects adjacent plant cells.
Chloroplast: Provides energy through photosynthesis.
Cell Wall: Made of cellulose. Provides structure to plants.
Animal
Vacuole: Smaller in animal cells. Stores water, nutrients, and waste products within the cell.
Extracellular Matrix: Helps with cell communication, growth, and movement.
Vesicle: Transports materials within the cell.
Peroxisome: Break down fatty acids and amino acids.
Cytoskeleton: Maintains cell structure.
Intermediate Filament: Anchors nucleus and maintains cell shape. Made of proteins from the keratin family.
Microfilament: Maintains cell shape, movement, and aids in muscle contraction. Made of actin protein.
Microtubule: Maintains cell shape and cell/ organelle movement. Made of tubulin protein.
Cytoplasm: Protects genetic material and cellular structures.
Ribosome: Synthesizes proteins.
Golgi Apparatus: Process and package proteins and lipid molecules.
Lysosome: Break down and recycle cellular structures.
Rough Endoplasmic Reticulum: Has bound ribosomes for protein synthesis.
Smooth Endoplasmic Reticulum: Synthesizes lipids and breaks down toxins.
Cell Membrane: Regulates the transport of materials entering and exiting the cell.
Cell Junctions: Connections between cells.
Gap: Anything can pass through.
Desmosomes: Some stuff can get through.
Tight: Nothing can pass through.
Mitochondria: Powerhouse of the cell. Produces ATP.
Nucleus: Holds genetic information such as DNA and RNA.
Nuclear Envelope: Regulates transportation of molecules through nuclear pores.