#2 ELONGATION: The RNA polymerase moves downstream towards transcription, unwinding the DNA and elongating the RNA transcript in a 5' to 3' direction.
RNA polymerase moves along the DNA untwisting the double helix and exposing approximately 10-20 DNA nucleotides at a time to pair it with RNA nucleotides.
#3 TERMINATION: RNA transcript is released and the RNA polymerase detaches from the DNA.
In Eukaryotes the RNA polymerase II transcribes a polyadenylation signal sequence.
This specifies a polyadenylation signal (AAUAAA) into the pre-mRNA bounding it by specific proteins in the nucleus.
AAUAAA cut the RNA transcript free from the polymerase and releases the pre-mRNA.
RNA Processing
The ends of the pre-mRNA are modified. The 5' end is synthesized first and receives a 5' cap. The 5' cap is a modified form of a guanine nucleotide.
Polyadenylate polymerase adds 50-250 Adenine nucleotides to the 3' end of the pre-mRNA to make a poly-A tail.
In Prokaryotes a RNA sequence called the terminator signals the end of transcription.
While transcription is occurring, DNA strands are re-forming a double helix.
Transcription in Prokaryotes
In cytoplasm
In Prokaryotes, the RNA polymerase recognizes where to start.
In nucleus
In a Eukaryotic promoter there is a TATA box: A nucleotide sequence that contains approximately 25 TATA nucleotides upstream from the starting point in the promoter.
Transcription factors (proteins) recognize the TATA box and bind to the DNA strand prior to the RNA polymerase II binds.
Additional transcription factors bind along to the RNA polymerase II.
RNA polymerase binds to the promoter (upstream)of a gene, unwinding the DNA to use the strand going from 3' to 5' as the template strand.
Transcription Initiation Complex: when transcription factors and RNA polymerase become bound to the promoter.
Direction of transcription = downstream The other direction = upstream
#1 INITIATION: RNA polymerase initiates RNA synthesis at the starting point on the template strand.
Transcription in Eukaryotes
Sites where Transcription Occurs
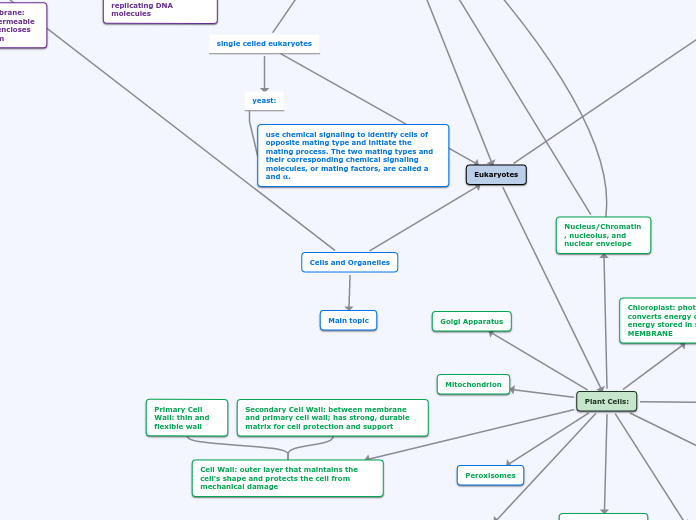
single celled eukaryotes
yeast:
use chemical signaling to identify cells of opposite mating type and initiate the mating process. The two mating types and their corresponding chemical signaling molecules, or mating factors, are called a and 𝛂.
Cell Signaling
Cells and Organelles
Main topic
Eukaryotes
Animal Cells:
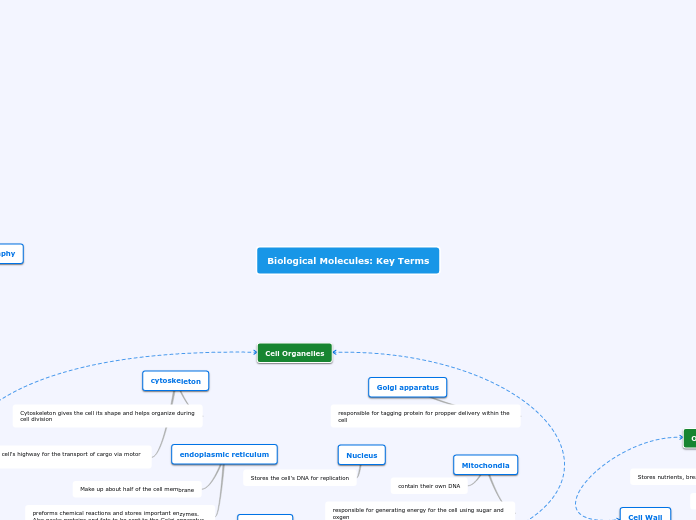
Endoplasmic Reticulum (ER): a network of membranous sacs; the ER membrane separates the internal compartment of the ER
Smooth ER: outer surface, lacks ribosomes, functions in metabolic processes (synthesis of lipids, metabolism of carbohydrates, detoxifies drugs and poisons, breaks down hormones
Rough ER: studded with ribosomes on the outer surface of the membrane, makes secretory proteins
Nucleus: contains most of the DNA; DOUBLE MEMBRANE
Chromatin: material consisting of DNA and proteins; visible in cell division as individual condensed chromosomes
Nucleolus: nonmembranous structure involved in production of ribosomes
Nuclear envelope: DOUBLE MEMBRANE enclosing the nucleus, separating its contents from the cytoplasm, pierce by nuclear pores that serve as "holes" that regulate entry and exit of materials through envelope; continuous with ER
Ribosomes: complexes that synthesize proteins; NO MEMBRANE
Bound Ribosomes: attached to the outside of the ER or nuclear envelope & make proteins for insertion into membranes
Free Ribosomes: suspended in the cytosol, proteins of these function within cytosol
Golgi Apparatus: organelle active in synthesis, modification, sorting, and secretion of cell products
Trans face: shipping department of Golgi, gives rise to vesicles that pinch off and travel to other sites
Cis face: receiving department of Golgi, located near the ER, transport vesicles used to more from ER to Golgi, ER lumen and membrane fuse with the Golgi membrane
products of the ER are modified, stored, and then sent to other destinations
Extracellular Matrix: helps cells to bind together and regulates several cell functions; made out of glycoproteins and carbohydrates.
Cells attach to the matrix with the help of fibronectin- binds to cell-surface receptor proteins (integrins) in the membrane
Contains collagen (glycoprotein) that forms strong fibers outside the cells
Centrosome: region where the cell's microtubules are initiates; contains a pair of centrioles
Flagellum: motility structure present in some animal cells; composed of microtubules
Cytoskeleton: reinforces cell shape, functions in cell movement, and components are made of protein; located in cytoplasm
Microfilaments: thin solid rods that are built from actin molecules; help maintain cell shape, changes in cell shape, muscle contraction, cytoplasmic streaming, cell motility, and cell division
Actin-protein interactions help cytoplasmic streaming (the circular flow of cytoplasm within cell)
Actin filaments and Myosin (motor protein) interact to cause contraction of muscle cells
Intermediate filaments: maintain cell shape, anchorage of organelles, and help with formation of nuclear lamina
Microtubules: hollow rods constructed from globular proteins called tubulins; maintain cell shape, cell motility in cilia and flagella, chromosome movements in cell division, and organelle movements
Dyneins have two "feet" that "walk" along the micotubules and use ATP to cause bending movement
Large motor proteins called dyneins are attached along each outer microtubule double and are responsible for the bending movement of flagella and cilia
Microvilli: projections that increase the cell's surface area
Peroxisomes: produces hydrogen peroxide as a by-product and then converts it to water; single-membrane organelle
Mitochondrion: organelle where cellular respiration occurs and most ATP is generated; double-membrane organelle
Inner membrane: coiled with infoldings (cristae) and divides the mitochondrion into TWO internal compartments
Mitochondrial Matrix: enclosed by the inner membrane, contains different enzymes and mitochondrial DNA and ribosomes
Intermembrane Space: narrow region between the inner and outer membrane
Outer membrane is SMOOTH
Lysosomes: digestive organelle where macromolecules are hydrolyzed (add water molecule); has hydrolytic enzymes and are single-membrane organelles
Autophagy: lysosomes fuse with vesicle containing damaged organelles and hydrolytic enzymes are used to recycle the cell's own organic material
Phagocytosis: lysosomes fuse with a food vacuole and hydrolytic enzymes digest the food particles
excessive leakage from A LOT of lysosomes leads to self-digestion of cell
Plant Cells:
Ribosomes
Rough and Smooth ER
Nucleus/Chromatin, nucleolus, and nuclear envelope
Sites where DNA Replication Occurs
Each cell has an ORI (origin of replication) where DNA replication begins; prokaryotes have one ORI and eukaryotes have many
1) Helicase: enzyme that untwists the 2 strands at the replication fork
2) Single-stranded Binding Proteins: bind to the unpaired DNA strands and keep them from re-pairing
3) Primase: enzyme that synthesizes a complementary RNA primer using the parent strand as the template; new DNA strand will start at the 3' end of the primer
4) DNA Polymerase III: enzyme that adds a DNA nucleotide to the 3' end of the primer and continues to add complementary DNA nucleotides to the template strand
5) DNA Polymerase I: removes RNA nucleotides of primer from 5' end and replaces them with DNA nucleotides
6) DNA Ligase: joins final nucleotide of DNA replacement to the first DNA nucleotide of the adjacent Okazaki fragment
Nuclease: a DNA-cutting enzyme that cuts a segment of DNA strand that contains damage; the gaps are filled with nucleotides using undamaged strand
Sliding clamp: converts DNA Poly. III from being distributive (falling off) to processive (staying on)
Replicates the leading strand 5' to 3' direction; continously; only one primer
Replicates the lagging strand away from replication fork, 5' to 3' direction, and discontinously synthesized in a series of Okazaki fragments, who each need a primer
Adds each monomer through dehydration reactions; is able to proofread its own work
Needs a primer & can only replicate 5' to 3' end
Topoisomerase: enzyme that helps relieve the strain by breaking, swiveling, and rejoining DNA strands
Subtopic
Golgi Apparatus
Mitochondrion
Peroxisomes
Cytoskeleton: only microfilaments and microtubules
Central Vacuole: functions include storage, breakdown of waste products, hydrolyze macromolecules, and plant growth
Plasmodesmata: cytoplasmic channels through cell walls that connect the cytoplasms of adjacent cells
Chloroplast: photosynthetic organelle; converts energy of sunlight to chemical energy stored in sugar molecules; DOUBLE MEMBRANE
Stroma: fluid outside the thylakoid, contains the chloroplast DNA, ribosomes, and enzymes
Membranes of the chloroplast divide the chloroplast into 3 compartments: intermembrane space, stroma, and thylakoid space
Thylakoids: another membranous system inside chloroplasts in the form of flattened, interconnected sacs; thylakoids are stacked; each stack is called a granum
Cell Wall: outer layer that maintains the cell's shape and protects the cell from mechanical damage
Secondary Cell Wall: between membrane and primary cell wall; has strong, durable matrix for cell protection and support
Primary Cell Wall: thin and flexible wall
Prokaryotes
Cytoplasm: interior of the cell
Cytosol: semifluid, jelly-like substance where cellular components are suspended
Peptidoglycan: located in the cell walls of BACTERIA CELLS and provides rigid mechanical support and anchors molecules
Endospores: help the cell survive under harsh environmental conditions i.e. lacking water or essential nutrients
Pili: appendages that help attach 2 cells together and used for bacterial mating
Flagella: organelle used for movement
Glycocalyx: outer coating of many prokaryotes, consisting of a capsule or slime layer
Slime layer: unorganized layer of extracellular fluid that protects the cell against dehydration (and other dangers) and helps it adhere to surfaces
Capsule: sticky layer of polysaccharides or proteins that protects the cell, helps maintain moisture, and helps the cell adhere to surfaces
Plasma membrane: selectively permeable barrier that encloses the cytoplasm
Ribosomes: complexes that synthesize proteins
Nucleoid: region where DNA is at (NO MEMBRANE); have a circular chromosome
Plasmids: smaller rings of independent replicating DNA molecules
Fimbrie: attachment structures on the surface of some prokaryotes; also used for bacterial mating
Cell wall: rigid structure outside of the plasma membrane that gives the cell shape