SQBU3723 (01)
Biocomputation and Bioinformatics
Assignment 2: Nucleic Acid Research (NAR) Database
Date: 6-3-2014
Group 7
CHAN ZHI XIN A11QB0054
LAI CHONG PAU A11QB0073
LOO FONG FONG A11QB0014
LOW HUI XIEN A11QB0082
NG NAN SEE A11QB0001
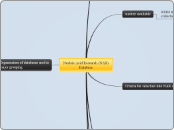
Nucleic Acid Research (NAR) Database
Organisation of databases and its major grouping
Cell Biology
Eg. NCBI Bookshelf
Eg. MethylomeDB
Eg. ExoCarta
Eg. CloneDB
Immunological Databases
Eg. The Immune Epitope Database (IEDB
Eg. Protegen
Eg. Epitome
Eg. AntigenDB
Plant Databases
Eg. Chloroplast Genome Database
Other plants
Rice
Arabidopsis thaliana
General plant databases
Eg. MetaCrop
Eg. GeneFarm
Organelle Databases
Eg. Plant Organelles Databases
Eg. Organelle genomes
Eg. Organelle DB
Mitochondrial genes and proteins
Eg. MitoGenesisDB
Eg. MitoDrome
Eg. Human MtDB
Other Molecular Biology Databases
Eg. PubMed
Eg. BioModels
Molecular probes and primers
RTPrimerDB
probeBase
PrimerBank
Human OligoGenome Resource
Drugs and drug design
Eg. SuperNatural
Eg. BacMet
Proteomics Databases
Eg. PeptideAtlas
Eg. 2D-PAGE
Microarray Data andother Gene Expression Databases
Eg. GeneTrap
Eg. GeneNote
Eg:Gene Expression Barcode
Eg. CycleBase
Human Genes and Diseases
Gene-, system-, disease-specific databases
Cancer gene databases
General polymorphism databases
General human genetics databases
Human and other Vertebrate Genomes
Human ORFs
Eg. PReMod
Eg. PlasmID
Eg. GeneSpeed
Human genome databses, maps and viewers
Eg. GeneLoc
Eg. GeneAnnot
Model organisms, comparative genomics
Eg. TreeFam
Eg. Mouse Phenome Database
Eg. Animal Genome Size Database
Metabolic and Signaling Pathways
Signalling pathway
Eg. SPIKE
Eg. Quorumpeps
Eg. Networkin
Protein-protein interaction
Eg. SynSysNet
Eg. VirusMINT
Eg. GeneNet
Eg. EndoNet
Metabolic pathwaqys
Eg. Reactom
Eg. Bionemo
Enzymes and enzymes nomenclature
Eg. MultiTaskDB
Eg. FunTree
Eg. NMPDR-National Microbial pathogen Data Resource
Genomics Databases (non-vertebrate)
Invertebrate genome databases
Eg. NEMBASE
Fungal genome databases
Eg. YEASTRACT
Eg. YeastNet
Unicellular eukaryotes genome databases
Eg. TBestDB
Eg. Camparasite
Prokaryotic genome databases
Eg. MicroScope
Eg. AlterORF
Viral genome databases
Eg. HIV Drud Resistance Database
Eg. HepSeq
General genomics databases
Eg. GenoList
Eg. BacMap
Taxonomy and identification
Eg. MetaRef
Eg. GeneTrees
Genome annotation terms, ontologies ad nomenclature
Eg. MetaBase
Eg. IUPAC Nomenclature database
Eg. BioThesaurus
Structural Databases
Protein structure
Eg. SitesBase
Eg. IDEAL
Eg. DSDBASE
Nucleic acid structure
Eg. RNA FRABASE
Eg. 3DNALandscapes
Carbohydrates
Eg. Monosaccharide Browser
Eg. GlycoMapsDB
Small molecules
Eg. SuperToxic
Eg. SuperDrug
Eg. DrugBank
Eg. ChemBank
Protein Sequences Databases
Databases of individual protein families
Eg. SuperCYP
Eg. TransportDB
Eg. Histone Database
Eg. BACTIBASE
Protein domain databases; protein classification
Eg. OrthoDB
Eg. FunShift
Protein sequence motifs and active sites
Eg. PHOSIDA
Eg. eBLOCKS
Protein localization and targeting
Eg. PeroxisomeDB
Eg. CentrosomeDB
Eg. REFOLD
Eg. BindingDB
General sequence databases
Eg. UniProt
Eg. Patome
RNA Sequences Databases
The Small Subunit rRNA Modification Database
RNA Modification Database
Ribosomal Database Project (RDP-II)
HIV Sequence Database
Database for Bacterial Group II Introns
5S Ribosomal RNA Database
3D rRNA modification maps
16S and 23S Ribosomal RNA Mutation Database
Nucleotide Sequence Databases
Transcriptionnal regulator sites and transcription factors
Eg. TFClass
Eg. QuadBase
Gene structure, introns and exons, splice sites
Eg. Spliceosome Database
Eg. GeneTack
Coding and non-coding DNA
Eg. TranspoGene
Eg. Plant repeat database
Eg. MethDB
International Nucleotide Sequence Database Collaboration
Eg. The Sequence Read Archive (SRA)
Eg. NCBI Biosample/ BioProject
Eg. GenBank
Eg. DDBJ-DNA Data Bank of Japan
Why some databases are no longer in the the databases and dropped from it?
Limited budjets
Taken commercial route and no free version
Redundancy
Obsolete and non-responsive
Why databases are created and shared?
Ensure data available are up-to-date
As tools for genome analysis
To prevent loss of resources
For integration and cross-reference
Comparative genomics purposes between diverse organisms and species
Emphasis on genome data to improve human health and disease treatment
Why we need to group the databases?
To update the changes (deletion and insertion of databases)
To exploit the related resources or specific information easily and effectively
To track databases, methods and tools in specific field
To utilise the databases effectively when neccessary
To narrow down the search results
To manage an increasing number of informations in the systematic ways
Criteria for selection into NAR databases
Consideration of so-called ‘boutique’ databases, covering relatively narrow topics
Avoid accepting new EST databases
as these data have a home in the DDBJ, genbank and European Nucleotide Archive databases
particularly those dealing with individual species
Avoid accepting databases on gene expression
as the underlying data must be submitted to array express/ GEO
Supplement with convenient search tools and easy-to-use visualization
Providing a convenient one-stop source of disparate data not available elsewhere
Data warehouses, portals, cross-platform search tools and visualization tools
Freely available online as well as selected databases published elsewhere
Web-accessible databases that offer carefully curated data that are not available elsewhere
Degree of value added (usually in the form of manual curation) in the production of the database
Comprehensiveness of coverag
The general utility of the database to the scientific community
number available
online molecular biology database collection
1552 online databases
41 subcategories
15 major groups
The different types of databases that useful in searching information of haemoglobin
Protein sequence databases
Protein properties
BindingDB
3. For example, this database curates measured binding affinities for the association of haemoglobin with various small organic molecules and their inhibition effect respectively.
2. Provide a deep search for various detailed data related those assays.
1. From the full search results for haemoglobin, various haemoglobin-related assays are available.
General sequence database
NCBI Protein database
3. From the category of proteins, key in haemoglobin. The search results would show approximately 32656 records related to sequences for various haemoglobins, 898 records related to structures, 167 records associated with the sequence similarity-based protein clusters, followed by 16 records related to the conserved protein domains.
2. Search results for haemoglobin displayed various data types available and those data are nicely categorized into different categories such as literature, organisms, proteins, chemicals and pathways.
1. NCBI provides Global Cross-database Search.